Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
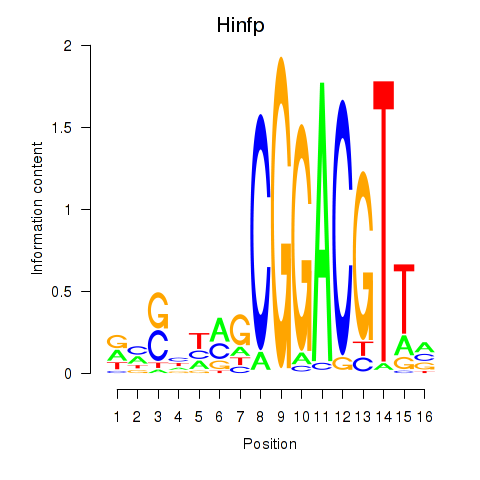
Results for Hinfp
Z-value: 0.56
Transcription factors associated with Hinfp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hinfp
|
ENSRNOG00000009499 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hinfp | rn6_v1_chr8_-_48634797_48634797 | -0.23 | 7.1e-01 | Click! |
Activity profile of Hinfp motif
Sorted Z-values of Hinfp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_221420271 | 0.23 |
ENSRNOT00000028481
|
LOC687780
|
similar to Finkel-Biskis-Reilly murine sarcoma virusubiquitously expressed |
| chr17_+_45175121 | 0.19 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr2_-_23289266 | 0.19 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr1_+_47942800 | 0.15 |
ENSRNOT00000079312
|
Wtap
|
Wilms tumor 1 associated protein |
| chr14_-_99780964 | 0.14 |
ENSRNOT00000006906
|
Sec61g
|
Sec61 translocon gamma subunit |
| chr3_+_65815080 | 0.14 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr10_-_94352880 | 0.14 |
ENSRNOT00000035973
|
Limd2
|
LIM domain containing 2 |
| chr8_-_55491152 | 0.14 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr1_+_141447022 | 0.13 |
ENSRNOT00000020690
|
AC096024.1
|
|
| chr2_-_116161998 | 0.13 |
ENSRNOT00000012560
|
Gpr160
|
G protein-coupled receptor 160 |
| chr5_-_76756140 | 0.12 |
ENSRNOT00000022107
ENSRNOT00000089251 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr16_+_36116258 | 0.11 |
ENSRNOT00000017652
|
Sap30
|
Sin3A associated protein 30 |
| chr9_-_100445158 | 0.11 |
ENSRNOT00000056558
|
Mterf4
|
mitochondrial transcription termination factor 4 |
| chr2_-_174012676 | 0.11 |
ENSRNOT00000013585
|
Pdcd10
|
programmed cell death 10 |
| chr1_-_82166363 | 0.10 |
ENSRNOT00000027774
|
Pafah1b3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 |
| chr2_+_115678344 | 0.10 |
ENSRNOT00000036717
|
Slc7a14
|
solute carrier family 7, member 14 |
| chrX_+_28435507 | 0.10 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr10_+_55626741 | 0.10 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr19_+_55300395 | 0.10 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr10_-_95934345 | 0.10 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr10_-_38969501 | 0.10 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr2_+_209161047 | 0.10 |
ENSRNOT00000023903
|
Dram2
|
DNA-damage regulated autophagy modulator 2 |
| chr17_-_21353134 | 0.10 |
ENSRNOT00000067898
|
Smim13
|
small integral membrane protein 13 |
| chrX_-_156440461 | 0.09 |
ENSRNOT00000083951
|
Rpl10
|
ribosomal protein L10 |
| chr18_-_27206777 | 0.09 |
ENSRNOT00000090569
|
AABR07031691.1
|
|
| chr20_+_34894419 | 0.09 |
ENSRNOT00000000471
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr5_+_141560192 | 0.09 |
ENSRNOT00000023354
|
Mycbp
|
Myc binding protein |
| chr5_-_58106706 | 0.09 |
ENSRNOT00000019561
|
Dctn3
|
dynactin subunit 3 |
| chr8_+_21663325 | 0.09 |
ENSRNOT00000027749
|
Ubl5
|
ubiquitin-like 5 |
| chr5_+_106952082 | 0.08 |
ENSRNOT00000046931
|
RGD1561246
|
similar to put. precursor MuIFN-alpha 5 |
| chr2_+_60949256 | 0.08 |
ENSRNOT00000025323
ENSRNOT00000040701 |
Amacr
|
alpha-methylacyl-CoA racemase |
| chr10_-_57653359 | 0.08 |
ENSRNOT00000089638
|
Derl2
|
derlin 2 |
| chr15_-_29548400 | 0.08 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr1_-_187766670 | 0.08 |
ENSRNOT00000092513
ENSRNOT00000092282 |
Rps15a
|
ribosomal protein S15a |
| chr8_+_111107358 | 0.08 |
ENSRNOT00000011203
|
Anapc13
|
anaphase promoting complex subunit 13 |
| chr1_+_219481575 | 0.08 |
ENSRNOT00000025507
|
Pold4
|
DNA polymerase delta 4, accessory subunit |
| chr13_+_96303703 | 0.08 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr16_+_49462889 | 0.08 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr8_+_70994563 | 0.08 |
ENSRNOT00000051504
ENSRNOT00000077163 |
Spg21
|
spastic paraplegia 21 homolog (human) |
| chr1_+_102900286 | 0.08 |
ENSRNOT00000017468
|
Ldha
|
lactate dehydrogenase A |
| chr5_+_172648950 | 0.08 |
ENSRNOT00000055361
|
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr6_+_123974798 | 0.08 |
ENSRNOT00000075794
|
Kcnk13
|
potassium two pore domain channel subfamily K member 13 |
| chr16_+_74292438 | 0.07 |
ENSRNOT00000026197
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr8_+_94243372 | 0.07 |
ENSRNOT00000012864
|
Rwdd2a
|
RWD domain containing 2A |
| chr5_+_14983371 | 0.07 |
ENSRNOT00000049609
|
Sumo4
|
small ubiquitin-like modifier 4 |
| chr17_+_27452638 | 0.07 |
ENSRNOT00000038349
|
Cage1
|
cancer antigen 1 |
| chr19_+_24846938 | 0.07 |
ENSRNOT00000045974
|
Ddx39a
|
DExD-box helicase 39A |
| chr1_+_154131926 | 0.07 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr4_+_181286455 | 0.07 |
ENSRNOT00000083633
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr15_-_20822740 | 0.07 |
ENSRNOT00000012957
|
Bmp4
|
bone morphogenetic protein 4 |
| chr2_+_147496229 | 0.07 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr19_+_39087990 | 0.07 |
ENSRNOT00000027553
|
Utp4
|
UTP4 small subunit processome component |
| chr2_+_140471690 | 0.07 |
ENSRNOT00000017379
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr5_+_105230580 | 0.07 |
ENSRNOT00000010056
|
Acer2
|
alkaline ceramidase 2 |
| chr10_-_56864005 | 0.07 |
ENSRNOT00000033098
ENSRNOT00000091131 |
Alox12
|
arachidonate 12-lipoxygenase, 12S type |
| chr6_+_124217241 | 0.07 |
ENSRNOT00000064679
|
Calm3
|
calmodulin 3 |
| chr10_+_72272248 | 0.07 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr2_-_28003186 | 0.07 |
ENSRNOT00000048554
|
Hexb
|
hexosaminidase subunit beta |
| chr6_-_107461041 | 0.07 |
ENSRNOT00000058127
|
Heatr4
|
HEAT repeat containing 4 |
| chr10_+_11879418 | 0.07 |
ENSRNOT00000068577
|
RGD1561796
|
RGD1561796 |
| chr1_+_201950144 | 0.07 |
ENSRNOT00000027967
|
Pstk
|
phosphoseryl-tRNA kinase |
| chr1_+_80383050 | 0.07 |
ENSRNOT00000023451
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr18_-_28438654 | 0.07 |
ENSRNOT00000036301
|
Mzb1
|
marginal zone B and B1 cell-specific protein |
| chr6_+_33786627 | 0.07 |
ENSRNOT00000008359
|
Pum2
|
pumilio RNA-binding family member 2 |
| chr4_-_27755103 | 0.07 |
ENSRNOT00000012253
|
Fam133b
|
family with sequence similarity 133, member B |
| chr10_+_109786222 | 0.07 |
ENSRNOT00000054953
|
Npb
|
neuropeptide B |
| chr14_+_104475082 | 0.06 |
ENSRNOT00000084481
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chr5_-_107388182 | 0.06 |
ENSRNOT00000058358
|
LOC100912356
|
interferon alpha-1-like |
| chr15_-_46432965 | 0.06 |
ENSRNOT00000014320
|
Gata4
|
GATA binding protein 4 |
| chr18_+_57654290 | 0.06 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr8_+_63119395 | 0.06 |
ENSRNOT00000046518
|
LOC685963
|
similar to 60S ribosomal protein L38 |
| chr4_-_81968832 | 0.06 |
ENSRNOT00000016608
|
Skap2
|
src kinase associated phosphoprotein 2 |
| chr17_+_10066187 | 0.06 |
ENSRNOT00000091200
|
Uimc1
|
ubiquitin interaction motif containing 1 |
| chr12_-_19114399 | 0.06 |
ENSRNOT00000073099
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr3_-_148010176 | 0.06 |
ENSRNOT00000031683
|
Defb29
|
defensin beta 29 |
| chr1_-_64175099 | 0.06 |
ENSRNOT00000091288
|
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr3_-_176144531 | 0.06 |
ENSRNOT00000082266
|
Tcfl5
|
transcription factor like 5 |
| chr1_+_222907159 | 0.06 |
ENSRNOT00000032123
|
Hrasls5
|
HRAS-like suppressor family, member 5 |
| chr1_+_212281237 | 0.06 |
ENSRNOT00000075049
|
Utf1
|
undifferentiated embryonic cell transcription factor 1 |
| chr1_+_205842489 | 0.06 |
ENSRNOT00000081610
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chrX_+_156355376 | 0.06 |
ENSRNOT00000078304
|
Lage3
|
L antigen family, member 3 |
| chr5_-_150609352 | 0.06 |
ENSRNOT00000074374
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr12_+_13090367 | 0.06 |
ENSRNOT00000001417
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr7_-_12283608 | 0.06 |
ENSRNOT00000029791
|
Rps15
|
ribosomal protein S15 |
| chr15_+_33069382 | 0.06 |
ENSRNOT00000060193
|
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr10_+_109630099 | 0.06 |
ENSRNOT00000072099
|
Ccdc137
|
coiled-coil domain containing 137 |
| chr2_+_149213134 | 0.06 |
ENSRNOT00000014470
ENSRNOT00000083504 |
Med12l
|
mediator complex subunit 12-like |
| chr1_-_221420115 | 0.06 |
ENSRNOT00000028475
|
Mrpl49
|
mitochondrial ribosomal protein L49 |
| chr5_+_101526551 | 0.06 |
ENSRNOT00000015254
|
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr14_+_115166416 | 0.06 |
ENSRNOT00000088916
ENSRNOT00000078329 |
Psme4
|
proteasome activator subunit 4 |
| chr13_-_110257367 | 0.06 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr19_+_26173771 | 0.06 |
ENSRNOT00000005581
|
Fbxw9
|
F-box and WD repeat domain containing 9 |
| chr2_+_23289374 | 0.06 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr17_-_78735324 | 0.06 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_+_141488272 | 0.06 |
ENSRNOT00000034042
|
Wdr93
|
WD repeat domain 93 |
| chr20_-_5076539 | 0.05 |
ENSRNOT00000076253
ENSRNOT00000037613 |
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr6_+_10642700 | 0.05 |
ENSRNOT00000040329
|
LOC100359600
|
karyopherin alpha 2-like |
| chr3_+_5519990 | 0.05 |
ENSRNOT00000070873
ENSRNOT00000007640 |
Adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr13_-_113817995 | 0.05 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chr20_+_34895059 | 0.05 |
ENSRNOT00000084420
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr6_-_50923510 | 0.05 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr5_-_105212173 | 0.05 |
ENSRNOT00000010383
|
Rps6
|
ribosomal protein S6 |
| chr7_+_11784341 | 0.05 |
ENSRNOT00000026155
|
Plekhj1
|
pleckstrin homology domain containing J1 |
| chr4_-_77510202 | 0.05 |
ENSRNOT00000038550
|
Zfp786
|
zinc finger protein 786 |
| chr4_+_118243132 | 0.05 |
ENSRNOT00000023654
|
RGD1306746
|
similar to Hypothetical protein MGC25529 |
| chr1_+_47942500 | 0.05 |
ENSRNOT00000025936
|
Wtap
|
Wilms tumor 1 associated protein |
| chr6_+_28663602 | 0.05 |
ENSRNOT00000005402
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr1_-_55895636 | 0.05 |
ENSRNOT00000029537
|
Vom2r9
|
vomeronasal 2 receptor, 9 |
| chr1_+_84293102 | 0.05 |
ENSRNOT00000028468
|
Sertad1
|
SERTA domain containing 1 |
| chr20_+_46168177 | 0.05 |
ENSRNOT00000057175
|
Zbtb24
|
zinc finger and BTB domain containing 24 |
| chr4_-_157294047 | 0.05 |
ENSRNOT00000005601
|
Eno2
|
enolase 2 |
| chr17_-_1648973 | 0.05 |
ENSRNOT00000036212
|
Slc35d2
|
solute carrier family 35 member D2 |
| chr2_+_30664217 | 0.05 |
ENSRNOT00000076786
|
Ak6
|
adenylate kinase 6 |
| chr20_-_4368693 | 0.05 |
ENSRNOT00000000501
|
Rnf5
|
ring finger protein 5 |
| chr1_-_198839228 | 0.05 |
ENSRNOT00000052100
|
LOC102553866
|
zinc finger protein 764-like |
| chr18_+_63394900 | 0.05 |
ENSRNOT00000024126
|
Psmg2
|
proteasome assembly chaperone 2 |
| chr7_-_51353068 | 0.05 |
ENSRNOT00000008222
|
Pawr
|
pro-apoptotic WT1 regulator |
| chr20_-_45259928 | 0.05 |
ENSRNOT00000087226
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr7_+_117351648 | 0.05 |
ENSRNOT00000056421
|
Smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr1_+_60717386 | 0.05 |
ENSRNOT00000015019
|
Ppp2r1a
|
protein phosphatase 2 scaffold subunit A alpha |
| chr15_+_51222954 | 0.05 |
ENSRNOT00000064428
ENSRNOT00000088928 |
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr2_+_113066885 | 0.05 |
ENSRNOT00000034960
|
Ghsr
|
growth hormone secretagogue receptor |
| chr17_-_52477575 | 0.05 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr14_-_104612597 | 0.05 |
ENSRNOT00000007002
|
Slc1a4
|
solute carrier family 1 member 4 |
| chr16_+_2338333 | 0.05 |
ENSRNOT00000017692
|
Arf4
|
ADP-ribosylation factor 4 |
| chr6_+_2969333 | 0.05 |
ENSRNOT00000047356
|
Morn2
|
MORN repeat containing 2 |
| chr3_-_154716239 | 0.05 |
ENSRNOT00000019453
|
RGD1563354
|
similar to hypothetical protein D630003M21 |
| chr1_-_157700064 | 0.05 |
ENSRNOT00000031974
|
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr1_-_276309099 | 0.05 |
ENSRNOT00000087680
|
Zdhhc6
|
zinc finger, DHHC-type containing 6 |
| chr14_+_113530470 | 0.05 |
ENSRNOT00000004919
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr1_+_46978458 | 0.05 |
ENSRNOT00000024211
|
Gtf2h5
|
general transcription factor IIH subunit 5 |
| chr1_-_220746224 | 0.05 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr2_+_236233239 | 0.05 |
ENSRNOT00000013694
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr7_+_136037230 | 0.05 |
ENSRNOT00000071423
|
AABR07058745.1
|
|
| chr9_+_98313632 | 0.05 |
ENSRNOT00000027012
|
Ramp1
|
receptor activity modifying protein 1 |
| chr1_-_219144610 | 0.05 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr1_-_198120061 | 0.04 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr8_-_73673317 | 0.04 |
ENSRNOT00000043006
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr2_-_188252725 | 0.04 |
ENSRNOT00000027608
|
Dap3
|
death associated protein 3 |
| chr9_-_99818262 | 0.04 |
ENSRNOT00000056600
|
Cops9
|
COP9 signalosome subunit 9 |
| chr5_-_153708114 | 0.04 |
ENSRNOT00000056105
ENSRNOT00000082037 |
Srrm1
|
serine and arginine repetitive matrix 1 |
| chr13_+_80460890 | 0.04 |
ENSRNOT00000004243
|
Vamp4
|
vesicle-associated membrane protein 4 |
| chr6_-_132608600 | 0.04 |
ENSRNOT00000015855
|
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr18_-_29504975 | 0.04 |
ENSRNOT00000063841
ENSRNOT00000024313 |
Apbb3
|
amyloid beta precursor protein binding family B member 3 |
| chr13_-_100740728 | 0.04 |
ENSRNOT00000000074
|
Fbxo28
|
F-box protein 28 |
| chr20_+_6397022 | 0.04 |
ENSRNOT00000002553
|
Rab44
|
RAB44, member RAS oncogene family |
| chr8_-_130491998 | 0.04 |
ENSRNOT00000064114
|
Higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr4_+_61420009 | 0.04 |
ENSRNOT00000082175
ENSRNOT00000011692 |
Lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr20_-_5112872 | 0.04 |
ENSRNOT00000076983
ENSRNOT00000001123 ENSRNOT00000076528 |
Csnk2b
|
casein kinase 2 beta |
| chr10_+_110406450 | 0.04 |
ENSRNOT00000054924
|
Hexdc
|
hexosaminidase D |
| chr14_+_84368476 | 0.04 |
ENSRNOT00000006827
|
Rnf215
|
ring finger protein 215 |
| chr1_-_193459396 | 0.04 |
ENSRNOT00000020281
|
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr16_+_8302950 | 0.04 |
ENSRNOT00000066062
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr3_+_148438939 | 0.04 |
ENSRNOT00000064196
|
Ttll9
|
tubulin tyrosine ligase like 9 |
| chr5_-_169301728 | 0.04 |
ENSRNOT00000013646
|
Espn
|
espin |
| chr1_+_220746387 | 0.04 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr15_-_86142672 | 0.04 |
ENSRNOT00000057830
|
Commd6
|
COMM domain containing 6 |
| chr11_-_57993548 | 0.04 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr1_+_174132798 | 0.04 |
ENSRNOT00000019247
|
Rpl27a
|
ribosomal protein L27a |
| chr17_+_29638677 | 0.04 |
ENSRNOT00000068617
|
Rpp40
|
ribonuclease P/MRP 40 subunit |
| chr8_-_122604787 | 0.04 |
ENSRNOT00000047198
|
Trim71
|
tripartite motif containing 71 |
| chr14_+_18983853 | 0.04 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr1_-_214202853 | 0.04 |
ENSRNOT00000022954
|
Lmntd2
|
lamin tail domain containing 2 |
| chr20_-_4677034 | 0.04 |
ENSRNOT00000075244
|
Rn60_20_0047.3
|
|
| chr6_-_1942972 | 0.04 |
ENSRNOT00000048711
|
Cdc42ep3
|
CDC42 effector protein 3 |
| chrX_+_43626480 | 0.04 |
ENSRNOT00000068078
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr1_+_219077771 | 0.04 |
ENSRNOT00000022824
|
Chka
|
choline kinase alpha |
| chr10_+_10530365 | 0.04 |
ENSRNOT00000003843
|
Eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr8_-_23148396 | 0.04 |
ENSRNOT00000075237
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr1_+_240908483 | 0.04 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr9_-_92435363 | 0.04 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr10_-_77512032 | 0.04 |
ENSRNOT00000003295
|
Pctp
|
phosphatidylcholine transfer protein |
| chr7_+_24939498 | 0.04 |
ENSRNOT00000010601
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr19_-_37370429 | 0.04 |
ENSRNOT00000022497
|
Kctd19
|
potassium channel tetramerization domain containing 19 |
| chr2_-_188252551 | 0.04 |
ENSRNOT00000088327
|
Dap3
|
death associated protein 3 |
| chr10_+_56710965 | 0.04 |
ENSRNOT00000087121
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr1_-_176079125 | 0.04 |
ENSRNOT00000047044
|
RGD1566189
|
similar to ferritin light chain |
| chr12_+_19573909 | 0.04 |
ENSRNOT00000029279
|
Lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr15_-_44411004 | 0.04 |
ENSRNOT00000031163
|
Cdca2
|
cell division cycle associated 2 |
| chr8_-_22091450 | 0.04 |
ENSRNOT00000028127
|
Fdx1l
|
ferredoxin 1-like |
| chr5_-_169167831 | 0.04 |
ENSRNOT00000012407
|
Phf13
|
PHD finger protein 13 |
| chr1_-_146289465 | 0.04 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr5_+_33580944 | 0.04 |
ENSRNOT00000092054
ENSRNOT00000036050 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr17_+_42241159 | 0.03 |
ENSRNOT00000024878
|
Acot13
|
acyl-CoA thioesterase 13 |
| chr17_-_79676499 | 0.03 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chr15_+_4125373 | 0.03 |
ENSRNOT00000029615
|
Usp54
|
ubiquitin specific peptidase 54 |
| chr6_-_133716847 | 0.03 |
ENSRNOT00000072399
|
Rtl1
|
retrotransposon-like 1 |
| chr9_-_94601852 | 0.03 |
ENSRNOT00000022485
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr17_+_82065937 | 0.03 |
ENSRNOT00000051349
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr7_+_14486638 | 0.03 |
ENSRNOT00000007548
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39 |
| chr8_+_114986326 | 0.03 |
ENSRNOT00000038166
|
Poc1a
|
POC1 centriolar protein A |
| chr4_+_77212203 | 0.03 |
ENSRNOT00000085805
|
Cul1
|
cullin 1 |
| chr11_-_24199968 | 0.03 |
ENSRNOT00000074032
|
Mrpl39
|
mitochondrial ribosomal protein L39 |
| chr4_-_82186190 | 0.03 |
ENSRNOT00000071729
|
LOC100911622
|
homeobox protein Hox-A7-like |
| chr17_+_29639079 | 0.03 |
ENSRNOT00000021877
|
Rpp40
|
ribonuclease P/MRP 40 subunit |
| chr10_+_55555089 | 0.03 |
ENSRNOT00000006597
|
Slc25a35
|
solute carrier family 25, member 35 |
| chr7_+_143882000 | 0.03 |
ENSRNOT00000017110
ENSRNOT00000091053 |
Mfsd5
|
major facilitator superfamily domain containing 5 |
| chr13_+_49195325 | 0.03 |
ENSRNOT00000004810
|
Dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr9_+_69953440 | 0.03 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_+_24449485 | 0.03 |
ENSRNOT00000073688
|
Tbca
|
tubulin folding cofactor A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hinfp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:1903660 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.1 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:1902462 | intermediate mesoderm development(GO:0048389) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) cardiac jelly development(GO:1905072) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) negative regulation of connective tissue replacement(GO:1905204) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:2001274 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0035928 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:1903347 | oogenesis stage(GO:0022605) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0019661 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.0 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 0.0 | 0.0 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.0 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.0 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.0 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0099567 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |