Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
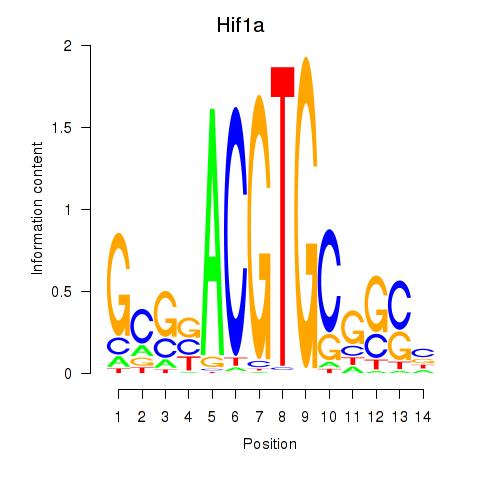
Results for Hif1a
Z-value: 0.83
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSRNOG00000008292 | hypoxia inducible factor 1 alpha subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hif1a | rn6_v1_chr6_+_96810907_96810907 | 0.74 | 1.5e-01 | Click! |
Activity profile of Hif1a motif
Sorted Z-values of Hif1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_4362717 | 0.92 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr7_-_13751271 | 0.91 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chr17_-_9695292 | 0.61 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr3_+_147585947 | 0.56 |
ENSRNOT00000006833
|
Scrt2
|
scratch family transcriptional repressor 2 |
| chr10_+_59529785 | 0.56 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr16_+_49462889 | 0.38 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr1_+_33910912 | 0.36 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr20_-_29511382 | 0.35 |
ENSRNOT00000085026
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr7_-_119441487 | 0.33 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr2_-_197991198 | 0.32 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_197991574 | 0.31 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr1_+_163445527 | 0.31 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr15_-_28406046 | 0.22 |
ENSRNOT00000015418
|
Zfp219
|
zinc finger protein 219 |
| chr20_-_13706205 | 0.21 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr10_-_104054805 | 0.20 |
ENSRNOT00000004853
|
Nt5c
|
5', 3'-nucleotidase, cytosolic |
| chr8_-_104221342 | 0.19 |
ENSRNOT00000015476
|
Atp1b3
|
ATPase Na+/K+ transporting subunit beta 3 |
| chr20_+_46429222 | 0.18 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr12_+_52359310 | 0.17 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr1_+_12823363 | 0.17 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr6_+_137997335 | 0.15 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr3_+_151032952 | 0.15 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr1_-_174378926 | 0.15 |
ENSRNOT00000018309
|
Tmem9b
|
TMEM9 domain family, member B |
| chr1_+_86873253 | 0.14 |
ENSRNOT00000071239
|
Fbxo27
|
F-box protein 27 |
| chr12_-_31323810 | 0.14 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr14_-_37770059 | 0.14 |
ENSRNOT00000029721
|
Slc10a4
|
solute carrier family 10, member 4 |
| chr11_+_84321078 | 0.14 |
ENSRNOT00000058043
|
Map6d1
|
MAP6 domain containing 1 |
| chr16_-_75855745 | 0.14 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr1_-_211265161 | 0.13 |
ENSRNOT00000080041
ENSRNOT00000023477 |
Bnip3
|
BCL2 interacting protein 3 |
| chr20_+_11393877 | 0.13 |
ENSRNOT00000001625
|
Pfkl
|
phosphofructokinase, liver type |
| chr17_-_27112820 | 0.12 |
ENSRNOT00000018359
|
Bmp6
|
bone morphogenetic protein 6 |
| chr2_-_23097610 | 0.11 |
ENSRNOT00000075286
|
Jmy
|
junction-mediating and regulatory protein |
| chr8_-_36314811 | 0.11 |
ENSRNOT00000013243
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr3_-_5976244 | 0.11 |
ENSRNOT00000009893
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr3_+_147643250 | 0.10 |
ENSRNOT00000000013
|
Tcf15
|
transcription factor 15 |
| chr11_+_80255790 | 0.10 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr2_-_84531192 | 0.10 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr20_+_11972381 | 0.09 |
ENSRNOT00000001642
|
Adarb1
|
adenosine deaminase, RNA-specific, B1 |
| chrX_+_106523278 | 0.08 |
ENSRNOT00000070802
|
MGC109340
|
similar to Microsomal signal peptidase 23 kDa subunit (SPase 22 kDa subunit) (SPC22/23) |
| chr10_-_29450644 | 0.08 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr5_+_147069616 | 0.08 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr4_-_157679962 | 0.07 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr7_-_97759852 | 0.07 |
ENSRNOT00000007484
|
Derl1
|
derlin 1 |
| chr3_-_5975734 | 0.07 |
ENSRNOT00000081376
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr3_-_176958880 | 0.07 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr16_+_9563218 | 0.07 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr1_-_146029840 | 0.07 |
ENSRNOT00000016455
|
Mesdc1
|
mesoderm development candidate 1 |
| chr16_+_19896986 | 0.07 |
ENSRNOT00000060355
|
Gtpbp3
|
GTP binding protein 3 |
| chr20_+_6018374 | 0.07 |
ENSRNOT00000000621
|
Mapk13
|
mitogen activated protein kinase 13 |
| chr14_+_61136746 | 0.07 |
ENSRNOT00000005126
|
Dhx15
|
DEAH-box helicase 15 |
| chr1_-_85220237 | 0.06 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr13_-_109629482 | 0.06 |
ENSRNOT00000072452
|
Flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr14_-_11198194 | 0.06 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr4_+_117235023 | 0.06 |
ENSRNOT00000021030
|
Cct7
|
chaperonin containing TCP1 subunit 7 |
| chr15_-_19655381 | 0.06 |
ENSRNOT00000009404
|
Ero1a
|
endoplasmic reticulum oxidoreductase 1 alpha |
| chr1_+_127802978 | 0.06 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr7_+_143810892 | 0.06 |
ENSRNOT00000016240
|
Znf740
|
zinc finger protein 740 |
| chr3_+_93495106 | 0.06 |
ENSRNOT00000029922
ENSRNOT00000085760 |
Abtb2
|
ankyrin repeat and BTB domain containing 2 |
| chr5_+_169506138 | 0.06 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr2_+_9526209 | 0.06 |
ENSRNOT00000021818
|
Lysmd3
|
LysM domain containing 3 |
| chr8_+_116325286 | 0.05 |
ENSRNOT00000017461
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr14_+_77322012 | 0.05 |
ENSRNOT00000088600
ENSRNOT00000041639 |
Zbtb49
|
zinc finger and BTB domain containing 49 |
| chr14_-_86333424 | 0.05 |
ENSRNOT00000083191
|
Nudcd3
|
NudC domain containing 3 |
| chr10_+_55687050 | 0.05 |
ENSRNOT00000057136
|
Per1
|
period circadian clock 1 |
| chr7_+_116943057 | 0.05 |
ENSRNOT00000056527
|
Tigd5
|
tigger transposable element derived 5 |
| chr4_-_117234928 | 0.05 |
ENSRNOT00000073135
|
Pradc1
|
protease-associated domain containing 1 |
| chr3_+_164822111 | 0.05 |
ENSRNOT00000014568
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr9_+_82033543 | 0.04 |
ENSRNOT00000023439
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr7_+_130431213 | 0.04 |
ENSRNOT00000055792
|
Mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr9_-_16612136 | 0.04 |
ENSRNOT00000023495
|
Mea1
|
male-enhanced antigen 1 |
| chr7_-_11824742 | 0.04 |
ENSRNOT00000051467
|
Dot1l
|
DOT1 like histone lysine methyltransferase |
| chr11_+_7210209 | 0.04 |
ENSRNOT00000076695
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr1_-_222178725 | 0.04 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr20_-_19825150 | 0.04 |
ENSRNOT00000032159
|
Ccdc6
|
coiled-coil domain containing 6 |
| chr5_-_78379346 | 0.04 |
ENSRNOT00000020625
|
Alad
|
aminolevulinate dehydratase |
| chr3_+_9643047 | 0.04 |
ENSRNOT00000035805
|
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr2_-_43393207 | 0.04 |
ENSRNOT00000017968
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr9_+_16612433 | 0.04 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr10_-_58973020 | 0.03 |
ENSRNOT00000020379
|
Smtnl2
|
smoothelin-like 2 |
| chr7_-_75422268 | 0.03 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr17_-_75886523 | 0.03 |
ENSRNOT00000046266
|
Usp6nl
|
USP6 N-terminal like |
| chr4_-_17594598 | 0.03 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr10_-_59049482 | 0.03 |
ENSRNOT00000078272
|
Spns2
|
spinster homolog 2 |
| chr1_+_219759183 | 0.03 |
ENSRNOT00000026316
|
Pc
|
pyruvate carboxylase |
| chr12_+_25093149 | 0.03 |
ENSRNOT00000050059
|
Eif4h
|
eukaryotic translation initiation factor 4H |
| chr12_+_39253409 | 0.03 |
ENSRNOT00000001774
|
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2 |
| chr1_+_165170645 | 0.03 |
ENSRNOT00000022879
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr4_+_29639154 | 0.03 |
ENSRNOT00000073687
|
Casd1
|
CAS1 domain containing 1 |
| chr1_+_102900286 | 0.03 |
ENSRNOT00000017468
|
Ldha
|
lactate dehydrogenase A |
| chr7_-_134560713 | 0.02 |
ENSRNOT00000006621
|
Yaf2
|
YY1 associated factor 2 |
| chr8_-_6203515 | 0.02 |
ENSRNOT00000087278
ENSRNOT00000031189 ENSRNOT00000008074 ENSRNOT00000085285 ENSRNOT00000007866 |
Yap1
|
yes-associated protein 1 |
| chr6_+_128750795 | 0.02 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr9_+_15068399 | 0.02 |
ENSRNOT00000064229
ENSRNOT00000087107 |
Foxp4
|
forkhead box P4 |
| chr19_-_55367353 | 0.02 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr4_+_140886545 | 0.02 |
ENSRNOT00000088273
|
Edem1
|
ER degradation enhancing alpha-mannosidase like protein 1 |
| chr9_-_61690956 | 0.02 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr9_-_16611899 | 0.02 |
ENSRNOT00000089168
|
Mea1
|
male-enhanced antigen 1 |
| chr14_+_60657686 | 0.01 |
ENSRNOT00000070892
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr2_-_148891900 | 0.01 |
ENSRNOT00000018488
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr8_+_114897011 | 0.01 |
ENSRNOT00000074683
|
Twf2
|
twinfilin actin-binding protein 2 |
| chr10_+_4411766 | 0.01 |
ENSRNOT00000003213
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr5_-_136748980 | 0.01 |
ENSRNOT00000026844
|
Ipo13
|
importin 13 |
| chr1_-_101457126 | 0.01 |
ENSRNOT00000087061
ENSRNOT00000028328 |
Bax
|
BCL2 associated X, apoptosis regulator |
| chr10_+_39435227 | 0.01 |
ENSRNOT00000042144
ENSRNOT00000077185 |
P4ha2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr1_-_91832058 | 0.01 |
ENSRNOT00000079088
|
Nudt19
|
nudix hydrolase 19 |
| chr14_+_83510278 | 0.00 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_+_103726238 | 0.00 |
ENSRNOT00000006776
|
Lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hif1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.0 | 0.2 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.0 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.1 | GO:0061744 | motor behavior(GO:0061744) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.4 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) regulation of memory T cell differentiation(GO:0043380) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |