Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hey2
Z-value: 0.28
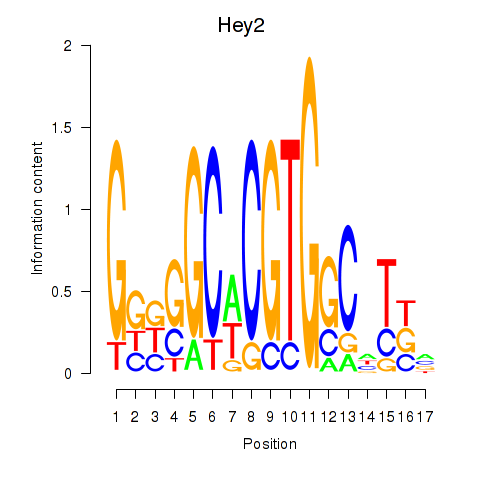
Transcription factors associated with Hey2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hey2
|
ENSRNOG00000013364 | hes-related family bHLH transcription factor with YRPW motif 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hey2 | rn6_v1_chr1_+_29191192_29191192 | -0.12 | 8.5e-01 | Click! |
Activity profile of Hey2 motif
Sorted Z-values of Hey2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_39711201 | 0.10 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr6_+_86785771 | 0.08 |
ENSRNOT00000066702
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr17_+_53229785 | 0.07 |
ENSRNOT00000082040
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr3_-_146470293 | 0.07 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr3_+_134413170 | 0.07 |
ENSRNOT00000074338
|
AABR07054000.1
|
|
| chr16_+_7103998 | 0.06 |
ENSRNOT00000016581
|
Pbrm1
|
polybromo 1 |
| chr3_-_51297852 | 0.06 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr6_+_73345392 | 0.06 |
ENSRNOT00000088378
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr1_+_11963836 | 0.05 |
ENSRNOT00000074325
|
AABR07000398.1
|
|
| chr6_-_92527711 | 0.05 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr13_-_76049363 | 0.05 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr20_-_3401273 | 0.05 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr8_+_72405748 | 0.05 |
ENSRNOT00000023952
|
Car12
|
carbonic anhydrase 12 |
| chr17_-_6244612 | 0.04 |
ENSRNOT00000042145
ENSRNOT00000090914 ENSRNOT00000082611 |
Ntrk2
|
neurotrophic receptor tyrosine kinase 2 |
| chr13_-_90832469 | 0.04 |
ENSRNOT00000086508
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr8_+_56179637 | 0.04 |
ENSRNOT00000035989
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr7_+_71036565 | 0.04 |
ENSRNOT00000005685
|
Zbtb39
|
zinc finger and BTB domain containing 39 |
| chr7_+_53878610 | 0.04 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr16_+_48120864 | 0.04 |
ENSRNOT00000068562
|
Stox2
|
storkhead box 2 |
| chr15_-_89339323 | 0.04 |
ENSRNOT00000013297
|
Rbm26
|
RNA binding motif protein 26 |
| chr6_-_109162267 | 0.04 |
ENSRNOT00000077518
|
Nek9
|
NIMA-related kinase 9 |
| chr10_-_61744976 | 0.03 |
ENSRNOT00000079926
ENSRNOT00000092314 ENSRNOT00000034298 |
Sgsm2
|
small G protein signaling modulator 2 |
| chr10_+_70136855 | 0.03 |
ENSRNOT00000084076
|
Lig3
|
DNA ligase 3 |
| chr10_+_62674561 | 0.03 |
ENSRNOT00000019946
ENSRNOT00000056110 |
Ankrd13b
|
ankyrin repeat domain 13B |
| chr5_-_25584278 | 0.03 |
ENSRNOT00000090579
ENSRNOT00000090376 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr13_-_70174565 | 0.03 |
ENSRNOT00000067135
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr16_-_7103604 | 0.03 |
ENSRNOT00000033677
|
Gnl3
|
G protein nucleolar 3 |
| chr8_-_47393503 | 0.03 |
ENSRNOT00000059868
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr2_+_207923775 | 0.03 |
ENSRNOT00000019997
ENSRNOT00000051835 |
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr15_-_89339037 | 0.03 |
ENSRNOT00000013255
|
Rbm26
|
RNA binding motif protein 26 |
| chr3_+_119698652 | 0.03 |
ENSRNOT00000088834
ENSRNOT00000066114 |
Stard7
|
StAR-related lipid transfer domain containing 7 |
| chr3_-_175479034 | 0.03 |
ENSRNOT00000086761
|
Hrh3
|
histamine receptor H3 |
| chr10_+_25890943 | 0.03 |
ENSRNOT00000086814
|
Nudcd2
|
NudC domain containing 2 |
| chr14_-_18720357 | 0.03 |
ENSRNOT00000047708
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr13_+_70174936 | 0.03 |
ENSRNOT00000064068
ENSRNOT00000079861 ENSRNOT00000092562 |
Arpc5
|
actin related protein 2/3 complex, subunit 5 |
| chr16_+_74177215 | 0.03 |
ENSRNOT00000025851
|
Ikbkb
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr1_+_256226179 | 0.03 |
ENSRNOT00000054748
|
Exoc6
|
exocyst complex component 6 |
| chr5_-_24255606 | 0.03 |
ENSRNOT00000037483
|
Plekhf2
|
pleckstrin homology and FYVE domain containing 2 |
| chr1_+_140602542 | 0.03 |
ENSRNOT00000085570
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr9_-_45505767 | 0.03 |
ENSRNOT00000033964
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr1_-_112947162 | 0.03 |
ENSRNOT00000014573
ENSRNOT00000083894 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr13_-_90832138 | 0.03 |
ENSRNOT00000010930
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr15_+_18399515 | 0.03 |
ENSRNOT00000071273
|
Fam107a
|
family with sequence similarity 107, member A |
| chr8_-_61079526 | 0.03 |
ENSRNOT00000068658
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr3_-_148722710 | 0.02 |
ENSRNOT00000090919
ENSRNOT00000068592 |
Plagl2
|
PLAG1 like zinc finger 2 |
| chr16_+_26906716 | 0.02 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr11_+_61661724 | 0.02 |
ENSRNOT00000079423
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr7_-_126913585 | 0.02 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr7_-_12673659 | 0.02 |
ENSRNOT00000091650
ENSRNOT00000041277 ENSRNOT00000044865 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr5_+_173340060 | 0.02 |
ENSRNOT00000067841
|
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr10_+_57462447 | 0.02 |
ENSRNOT00000041443
|
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr4_+_6559545 | 0.02 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr3_+_58084606 | 0.02 |
ENSRNOT00000084797
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr6_-_28464118 | 0.02 |
ENSRNOT00000068214
|
Efr3b
|
EFR3 homolog B |
| chr1_+_167373678 | 0.02 |
ENSRNOT00000027685
ENSRNOT00000088370 |
Stim1
|
stromal interaction molecule 1 |
| chr10_-_10725655 | 0.02 |
ENSRNOT00000061236
|
Ubn1
|
ubinuclein 1 |
| chr9_+_29984077 | 0.02 |
ENSRNOT00000075592
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 |
| chr15_-_108210826 | 0.02 |
ENSRNOT00000064705
|
Dock9
|
dedicator of cytokinesis 9 |
| chr7_+_27309966 | 0.02 |
ENSRNOT00000031871
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr1_-_220938814 | 0.02 |
ENSRNOT00000028081
|
Ovol1
|
ovo like transcriptional repressor 1 |
| chr14_-_80169431 | 0.02 |
ENSRNOT00000079769
ENSRNOT00000058315 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr14_-_10446909 | 0.02 |
ENSRNOT00000002962
|
Mrps18c
|
mitochondrial ribosomal protein S18C |
| chr1_+_167374182 | 0.02 |
ENSRNOT00000092882
|
Stim1
|
stromal interaction molecule 1 |
| chr1_+_86914137 | 0.02 |
ENSRNOT00000027006
|
Fbxo17
|
F-box protein 17 |
| chr8_+_116343096 | 0.02 |
ENSRNOT00000022092
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr20_+_50172618 | 0.02 |
ENSRNOT00000091456
|
Prep
|
prolyl endopeptidase |
| chr3_+_62648447 | 0.02 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr8_+_53295222 | 0.02 |
ENSRNOT00000009724
ENSRNOT00000067420 |
Usp28
|
ubiquitin specific peptidase 28 |
| chr4_-_98593664 | 0.02 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr15_+_40937708 | 0.02 |
ENSRNOT00000018407
|
Spata13
|
spermatogenesis associated 13 |
| chr5_+_135735825 | 0.02 |
ENSRNOT00000068267
|
Zswim5
|
zinc finger, SWIM-type containing 5 |
| chr2_-_46544457 | 0.02 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr4_+_151250041 | 0.02 |
ENSRNOT00000009832
|
Dcp1b
|
decapping mRNA 1B |
| chr3_-_137969658 | 0.02 |
ENSRNOT00000007727
|
Bfsp1
|
beaded filament structural protein 1 |
| chr1_-_280233755 | 0.02 |
ENSRNOT00000064463
|
Shtn1
|
shootin 1 |
| chr4_+_84770375 | 0.01 |
ENSRNOT00000013528
|
Plekha8
|
pleckstrin homology domain containing A8 |
| chr14_-_92117923 | 0.01 |
ENSRNOT00000044726
|
Grb10
|
growth factor receptor bound protein 10 |
| chr5_+_165405168 | 0.01 |
ENSRNOT00000014934
|
Srm
|
spermidine synthase |
| chr1_-_112947399 | 0.01 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr15_-_44627765 | 0.01 |
ENSRNOT00000058887
|
Dock5
|
dedicator of cytokinesis 5 |
| chr9_+_94880053 | 0.01 |
ENSRNOT00000024445
|
Atg16l1
|
autophagy related 16-like 1 |
| chr16_+_48121430 | 0.01 |
ENSRNOT00000090839
|
Stox2
|
storkhead box 2 |
| chr9_+_66495832 | 0.01 |
ENSRNOT00000022676
ENSRNOT00000091283 |
Nop58
|
NOP58 ribonucleoprotein |
| chr11_+_32211115 | 0.01 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr5_+_90419020 | 0.01 |
ENSRNOT00000058893
|
Rasef
|
RAS and EF hand domain containing |
| chr10_-_78219793 | 0.01 |
ENSRNOT00000003369
|
Tom1l1
|
target of myb1 like 1 membrane trafficking protein |
| chr3_-_156905892 | 0.01 |
ENSRNOT00000022424
|
Emilin3
|
elastin microfibril interfacer 3 |
| chr14_+_77712240 | 0.01 |
ENSRNOT00000009101
|
Msx1
|
msh homeobox 1 |
| chr3_-_92783569 | 0.01 |
ENSRNOT00000009073
ENSRNOT00000009000 |
Cd44
|
CD44 molecule (Indian blood group) |
| chr7_+_80351774 | 0.01 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
| chr8_+_40001030 | 0.01 |
ENSRNOT00000083778
|
Esam
|
endothelial cell adhesion molecule |
| chr1_-_141533908 | 0.01 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr17_-_15643489 | 0.01 |
ENSRNOT00000021022
|
Ippk
|
inositol-pentakisphosphate 2-kinase |
| chr16_-_21348391 | 0.01 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr7_+_77678968 | 0.01 |
ENSRNOT00000006917
|
Atp6v1c1
|
ATPase H+ transporting V1 subunit C1 |
| chr6_-_1622196 | 0.01 |
ENSRNOT00000007492
|
Prkd3
|
protein kinase D3 |
| chr7_-_97944491 | 0.01 |
ENSRNOT00000008454
|
Zhx1
|
zinc fingers and homeoboxes 1 |
| chr7_+_94795214 | 0.01 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr15_+_106257650 | 0.01 |
ENSRNOT00000014859
ENSRNOT00000072538 |
Ipo5
|
importin 5 |
| chr8_-_64800467 | 0.01 |
ENSRNOT00000073122
|
Nr2e3
|
nuclear receptor subfamily 2, group E, member 3 |
| chr15_-_33589522 | 0.01 |
ENSRNOT00000022514
|
Efs
|
embryonal Fyn-associated substrate |
| chr10_+_47961056 | 0.01 |
ENSRNOT00000027312
|
Fam83g
|
family with sequence similarity 83, member G |
| chr3_+_125503638 | 0.01 |
ENSRNOT00000028900
|
Crls1
|
cardiolipin synthase 1 |
| chr15_-_52029816 | 0.01 |
ENSRNOT00000013067
|
Slc39a14
|
solute carrier family 39 member 14 |
| chr3_+_175408629 | 0.01 |
ENSRNOT00000081344
|
Lsm14b
|
LSM family member 14B |
| chr10_+_10725819 | 0.01 |
ENSRNOT00000004159
|
Glyr1
|
glyoxylate reductase 1 homolog |
| chr19_-_44158621 | 0.01 |
ENSRNOT00000031243
ENSRNOT00000088297 |
Tmem231
|
transmembrane protein 231 |
| chr9_-_29337922 | 0.01 |
ENSRNOT00000018896
|
RGD1309049
|
similar to RIKEN cDNA 4933415F23 |
| chr16_+_73564243 | 0.01 |
ENSRNOT00000030960
|
Golga7
|
golgin A7 |
| chr7_-_11330278 | 0.01 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr15_+_45422010 | 0.01 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr3_-_80003032 | 0.01 |
ENSRNOT00000017360
|
Madd
|
MAP-kinase activating death domain |
| chr7_+_117763783 | 0.01 |
ENSRNOT00000021208
|
Mfsd3
|
major facilitator superfamily domain containing 3 |
| chr4_-_66624912 | 0.01 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr11_-_34791993 | 0.01 |
ENSRNOT00000002284
|
Dscr3
|
DSCR3 arrestin fold containing |
| chr1_-_200163106 | 0.00 |
ENSRNOT00000027644
|
Mcmbp
|
minichromosome maintenance complex binding protein |
| chr7_-_106653567 | 0.00 |
ENSRNOT00000042522
|
Oc90
|
otoconin 90 |
| chr5_-_160070916 | 0.00 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr13_+_83972212 | 0.00 |
ENSRNOT00000004394
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr15_+_58554374 | 0.00 |
ENSRNOT00000058206
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr7_-_60416925 | 0.00 |
ENSRNOT00000008193
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr1_+_113034227 | 0.00 |
ENSRNOT00000081831
ENSRNOT00000077877 ENSRNOT00000077594 |
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr3_+_164822111 | 0.00 |
ENSRNOT00000014568
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr10_-_81653717 | 0.00 |
ENSRNOT00000003611
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr9_-_82195722 | 0.00 |
ENSRNOT00000077085
ENSRNOT00000084276 |
Cfap65
|
cilia and flagella associated protein 65 |
| chr16_-_21347968 | 0.00 |
ENSRNOT00000068554
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr1_+_101682172 | 0.00 |
ENSRNOT00000028540
|
Car11
|
carbonic anhydrase 11 |
| chr10_+_61685241 | 0.00 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr7_-_117070936 | 0.00 |
ENSRNOT00000048446
|
Fam83h
|
family with sequence similarity 83, member H |
| chr19_+_11473541 | 0.00 |
ENSRNOT00000082066
|
Amfr
|
autocrine motility factor receptor |
| chr14_+_7169519 | 0.00 |
ENSRNOT00000003026
|
Klhl8
|
kelch-like family member 8 |
| chr17_-_15511972 | 0.00 |
ENSRNOT00000074041
|
Aspnl1
|
asporin-like 1 |
| chr1_-_125229052 | 0.00 |
ENSRNOT00000038511
|
Mphosph10
|
M-phase phosphoprotein 10 |
| chr5_+_2043583 | 0.00 |
ENSRNOT00000082813
|
Eloc
|
elongin C |
| chr17_+_33408722 | 0.00 |
ENSRNOT00000023691
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr11_-_87485833 | 0.00 |
ENSRNOT00000079948
|
AABR07034751.1
|
|
| chr5_-_58124681 | 0.00 |
ENSRNOT00000019795
|
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hey2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.0 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |