Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hey1_Myc_Mxi1
Z-value: 1.02
Transcription factors associated with Hey1_Myc_Mxi1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
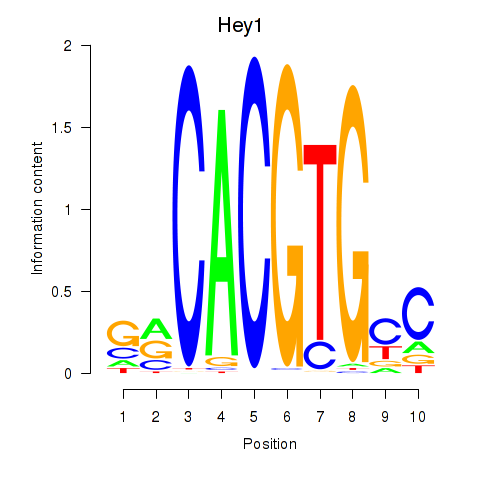
Hey1
|
ENSRNOG00000011593 | hes-related family bHLH transcription factor with YRPW motif 1 |
|
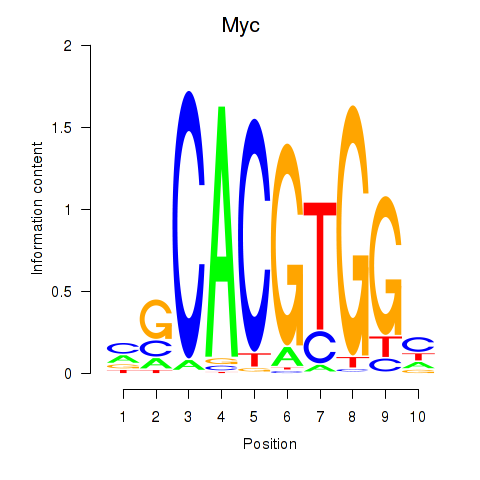
Myc
|
ENSRNOG00000004500 | myelocytomatosis oncogene |
|
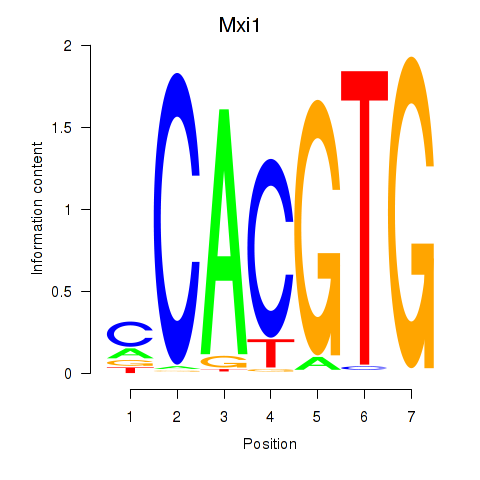
Mxi1
|
ENSRNOG00000034078 | MAX interactor 1, dimerization protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mxi1 | rn6_v1_chr1_+_274030978_274030978 | 0.60 | 2.9e-01 | Click! |
| Hey1 | rn6_v1_chr2_+_95320283_95320283 | -0.10 | 8.7e-01 | Click! |
| Myc | rn6_v1_chr7_+_102586313_102586313 | 0.06 | 9.3e-01 | Click! |
Activity profile of Hey1_Myc_Mxi1 motif
Sorted Z-values of Hey1_Myc_Mxi1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_123119460 | 2.14 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr2_-_197991198 | 1.61 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_197991574 | 1.52 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_187160373 | 0.87 |
ENSRNOT00000018961
|
Ntrk1
|
neurotrophic receptor tyrosine kinase 1 |
| chr2_-_260596777 | 0.80 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr17_-_9695292 | 0.77 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr14_+_4362717 | 0.62 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr4_+_33638709 | 0.58 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr16_+_49462889 | 0.57 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr3_+_113415774 | 0.57 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr13_-_90814119 | 0.47 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr5_-_147953093 | 0.47 |
ENSRNOT00000075270
|
Khdrbs1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr10_-_44746549 | 0.47 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr1_+_240908483 | 0.45 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr6_+_42852683 | 0.43 |
ENSRNOT00000079185
|
Odc1
|
ornithine decarboxylase 1 |
| chr1_+_73719005 | 0.42 |
ENSRNOT00000025110
|
Cdc42ep5
|
CDC42 effector protein 5 |
| chr10_+_91710495 | 0.40 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr4_-_165629996 | 0.40 |
ENSRNOT00000068185
ENSRNOT00000007427 |
Ybx3
|
Y box binding protein 3 |
| chr9_-_16848503 | 0.39 |
ENSRNOT00000024815
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr3_-_66417741 | 0.37 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_85574889 | 0.36 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr20_-_5533448 | 0.35 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr20_-_5533600 | 0.34 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr1_+_168964202 | 0.34 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr9_+_98490608 | 0.33 |
ENSRNOT00000027232
|
Klhl30
|
kelch-like family member 30 |
| chr17_-_13393243 | 0.32 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr3_+_147643250 | 0.30 |
ENSRNOT00000000013
|
Tcf15
|
transcription factor 15 |
| chr12_-_31323810 | 0.29 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr1_+_240776969 | 0.29 |
ENSRNOT00000049732
|
RGD1561453
|
similar to ribosomal protein S12 |
| chr10_-_81653717 | 0.29 |
ENSRNOT00000003611
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr6_+_137997335 | 0.29 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr3_-_120306551 | 0.28 |
ENSRNOT00000021082
|
Mall
|
mal, T-cell differentiation protein-like |
| chr3_+_61685619 | 0.28 |
ENSRNOT00000002140
|
Hoxd1
|
homeo box D1 |
| chr10_+_16542882 | 0.27 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr4_-_123118186 | 0.27 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr15_-_27849974 | 0.27 |
ENSRNOT00000012790
|
Osgep
|
O-sialoglycoprotein endopeptidase |
| chr1_-_168972725 | 0.26 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr12_-_23841049 | 0.26 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr7_+_128500011 | 0.26 |
ENSRNOT00000074625
|
Fam19a5
|
family with sequence similarity 19 member A5, C-C motif chemokine like |
| chr9_-_113331319 | 0.25 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chr1_-_214455039 | 0.25 |
ENSRNOT00000071028
|
Polr2l
|
RNA polymerase II subunit L |
| chrX_-_32355296 | 0.25 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr5_+_150080072 | 0.25 |
ENSRNOT00000047917
|
Tmem200b
|
transmembrane protein 200B |
| chr1_-_78212350 | 0.25 |
ENSRNOT00000071098
|
Inafm1
|
InaF-motif containing 1 |
| chr15_+_37790141 | 0.24 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr11_+_32211115 | 0.24 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr1_+_82452469 | 0.24 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr2_+_189400696 | 0.24 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr4_+_83391283 | 0.24 |
ENSRNOT00000031365
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr5_-_137372524 | 0.23 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr8_-_61917125 | 0.23 |
ENSRNOT00000085049
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr5_-_166726794 | 0.23 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr7_-_18683440 | 0.23 |
ENSRNOT00000068323
|
Rps28
|
ribosomal protein S28 |
| chr14_-_11198194 | 0.23 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr20_-_13706205 | 0.22 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr2_-_187668677 | 0.21 |
ENSRNOT00000056898
ENSRNOT00000092563 |
Tsacc
|
TSSK6 activating co-chaperone |
| chr9_-_97065817 | 0.21 |
ENSRNOT00000026419
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr1_-_265899958 | 0.21 |
ENSRNOT00000026013
|
Pitx3
|
paired-like homeodomain 3 |
| chr7_+_3332788 | 0.21 |
ENSRNOT00000010180
|
Cd63
|
Cd63 molecule |
| chr1_+_163445527 | 0.21 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr3_-_7498555 | 0.21 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr10_+_84135116 | 0.20 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr9_+_69953440 | 0.20 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr10_-_40375605 | 0.20 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr10_-_83898527 | 0.20 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr11_-_60613718 | 0.20 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr1_+_261229347 | 0.20 |
ENSRNOT00000018485
|
Ubtd1
|
ubiquitin domain containing 1 |
| chr10_+_83476107 | 0.19 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr11_-_34598102 | 0.19 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_-_199159125 | 0.19 |
ENSRNOT00000025618
|
Bcl7c
|
BCL tumor suppressor 7C |
| chr15_+_27849979 | 0.19 |
ENSRNOT00000013176
|
Apex1
|
apurinic/apyrimidinic endodeoxyribonuclease 1 |
| chr8_+_117117430 | 0.19 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr5_+_137371825 | 0.19 |
ENSRNOT00000072816
|
Tmem125
|
transmembrane protein 125 |
| chr1_-_101123402 | 0.19 |
ENSRNOT00000027976
|
Rpl13a
|
|
| chr7_-_11648322 | 0.19 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr1_+_12823363 | 0.19 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr3_-_81282157 | 0.19 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr3_-_8852192 | 0.18 |
ENSRNOT00000022786
|
Dolk
|
dolichol kinase |
| chr5_-_152464850 | 0.18 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr19_+_16415636 | 0.18 |
ENSRNOT00000089975
|
Irx5
|
iroquois homeobox 5 |
| chr7_+_141370491 | 0.18 |
ENSRNOT00000087662
|
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 |
| chr1_-_259357056 | 0.18 |
ENSRNOT00000022000
|
Pdlim1
|
PDZ and LIM domain 1 |
| chr4_-_129430251 | 0.18 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr1_+_80135391 | 0.18 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr10_+_16970626 | 0.18 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr6_+_136150785 | 0.17 |
ENSRNOT00000078211
ENSRNOT00000015308 |
LOC103692719
|
tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A |
| chr1_+_81230612 | 0.17 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr2_+_127538659 | 0.17 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr4_-_78879294 | 0.17 |
ENSRNOT00000084543
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr14_-_33580566 | 0.17 |
ENSRNOT00000063942
|
Paics
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr9_-_9702306 | 0.17 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr11_-_83985385 | 0.17 |
ENSRNOT00000002334
|
LOC102551435
|
endothelin-converting enzyme 2-like |
| chr10_+_55940533 | 0.17 |
ENSRNOT00000012061
|
RGD1563441
|
similar to RIKEN cDNA A030009H04 |
| chr5_-_166924136 | 0.17 |
ENSRNOT00000085251
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr4_+_7257752 | 0.17 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chrX_+_156863754 | 0.17 |
ENSRNOT00000083611
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr1_+_33910912 | 0.17 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr7_+_18683553 | 0.17 |
ENSRNOT00000009425
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr17_-_9234766 | 0.17 |
ENSRNOT00000015893
|
AABR07026997.1
|
|
| chr19_+_37226186 | 0.17 |
ENSRNOT00000075933
ENSRNOT00000065013 |
Hsf4
|
heat shock transcription factor 4 |
| chr17_+_85356042 | 0.17 |
ENSRNOT00000022201
|
Commd3
|
COMM domain containing 3 |
| chr5_-_157820889 | 0.16 |
ENSRNOT00000024326
|
Mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr10_-_81587858 | 0.16 |
ENSRNOT00000003582
|
Utp18
|
UTP18 small subunit processome component |
| chr12_+_12738812 | 0.16 |
ENSRNOT00000092233
ENSRNOT00000001379 |
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr2_+_206064394 | 0.16 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr15_-_34352673 | 0.16 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr8_-_66863476 | 0.16 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chrX_+_43497763 | 0.16 |
ENSRNOT00000005014
|
Prdx4
|
peroxiredoxin 4 |
| chr5_-_145423172 | 0.16 |
ENSRNOT00000081919
|
Gjb5
|
gap junction protein, beta 5 |
| chr1_+_220869805 | 0.16 |
ENSRNOT00000015962
|
Cfl1
|
cofilin 1 |
| chr5_+_150725654 | 0.15 |
ENSRNOT00000089852
ENSRNOT00000017740 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr10_-_57275708 | 0.15 |
ENSRNOT00000005370
|
Pfn1
|
profilin 1 |
| chr4_-_120817377 | 0.15 |
ENSRNOT00000021826
|
Podxl2
|
podocalyxin-like 2 |
| chr8_-_36314811 | 0.15 |
ENSRNOT00000013243
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr17_+_9837402 | 0.15 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr14_+_86673775 | 0.15 |
ENSRNOT00000091873
ENSRNOT00000079015 |
Ppia
|
peptidylprolyl isomerase A |
| chr5_+_169506138 | 0.15 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr1_-_82452281 | 0.15 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr10_+_55626741 | 0.15 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr6_-_137084739 | 0.15 |
ENSRNOT00000060943
|
Tmem179
|
transmembrane protein 179 |
| chr15_-_45376476 | 0.15 |
ENSRNOT00000065838
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr8_-_52814188 | 0.15 |
ENSRNOT00000087265
ENSRNOT00000047633 |
Rexo2
|
RNA exonuclease 2 |
| chr4_-_157679962 | 0.14 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_-_63499390 | 0.14 |
ENSRNOT00000032934
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr1_+_100393303 | 0.14 |
ENSRNOT00000026251
|
Syt3
|
synaptotagmin 3 |
| chrX_+_65226748 | 0.14 |
ENSRNOT00000076181
|
Msn
|
moesin |
| chr3_+_71020534 | 0.14 |
ENSRNOT00000007276
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr10_-_86509454 | 0.14 |
ENSRNOT00000009454
|
Ikzf3
|
IKAROS family zinc finger 3 |
| chr1_-_24191908 | 0.14 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_+_222229835 | 0.14 |
ENSRNOT00000051749
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr20_+_31265483 | 0.14 |
ENSRNOT00000079584
ENSRNOT00000000674 |
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr18_+_70996044 | 0.14 |
ENSRNOT00000024950
ENSRNOT00000066336 |
Dym
|
dymeclin |
| chr7_+_11724962 | 0.14 |
ENSRNOT00000026551
|
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_123494742 | 0.14 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr8_-_115140080 | 0.14 |
ENSRNOT00000015851
|
Acy1
|
aminoacylase 1 |
| chr1_-_78333321 | 0.14 |
ENSRNOT00000020402
|
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr1_+_40816107 | 0.14 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr17_-_9837293 | 0.13 |
ENSRNOT00000091320
ENSRNOT00000022134 |
Prelid1
|
PRELI domain containing 1 |
| chr3_-_173953684 | 0.13 |
ENSRNOT00000090468
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr6_-_26486695 | 0.13 |
ENSRNOT00000073236
|
Krtcap3
|
keratinocyte associated protein 3 |
| chr15_-_61564695 | 0.13 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr20_+_18493538 | 0.13 |
ENSRNOT00000000749
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr6_+_26797126 | 0.13 |
ENSRNOT00000010586
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr7_-_18634079 | 0.13 |
ENSRNOT00000010031
|
Angptl4
|
angiopoietin-like 4 |
| chr2_+_189106039 | 0.13 |
ENSRNOT00000028210
|
Ube2q1
|
ubiquitin conjugating enzyme E2 Q1 |
| chr1_-_164143818 | 0.13 |
ENSRNOT00000022557
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr3_+_177374812 | 0.13 |
ENSRNOT00000024096
|
Polr3k
|
RNA polymerase III subunit K |
| chr4_+_6827429 | 0.13 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr8_-_62386264 | 0.13 |
ENSRNOT00000026165
|
Cplx3
|
complexin 3 |
| chr19_+_14508616 | 0.13 |
ENSRNOT00000019192
|
Hmox1
|
heme oxygenase 1 |
| chr9_-_10054359 | 0.13 |
ENSRNOT00000072001
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr2_+_19823234 | 0.13 |
ENSRNOT00000022348
|
Rps23
|
ribosomal protein S23 |
| chr10_-_14022452 | 0.13 |
ENSRNOT00000016554
|
Npw
|
neuropeptide W |
| chr16_-_81706664 | 0.13 |
ENSRNOT00000026580
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr11_+_86890585 | 0.13 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr1_-_198706852 | 0.13 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr16_+_80826681 | 0.12 |
ENSRNOT00000000123
|
Coprs
|
coordinator of PRMT5 and differentiation stimulator |
| chr7_+_77966722 | 0.12 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr3_-_10375826 | 0.12 |
ENSRNOT00000093365
|
Ass1
|
argininosuccinate synthase 1 |
| chr19_-_57422093 | 0.12 |
ENSRNOT00000025512
|
RGD1559896
|
similar to RIKEN cDNA 2310022B05 |
| chr7_-_73270308 | 0.12 |
ENSRNOT00000007430
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr10_+_40208223 | 0.12 |
ENSRNOT00000000772
|
Hint1
|
histidine triad nucleotide binding protein 1 |
| chr1_-_226255886 | 0.12 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr1_-_89358166 | 0.12 |
ENSRNOT00000044532
|
Mag
|
myelin-associated glycoprotein |
| chr7_+_72772440 | 0.12 |
ENSRNOT00000009989
|
Mtdh
|
metadherin |
| chr1_-_91042230 | 0.12 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr7_-_140770647 | 0.12 |
ENSRNOT00000081784
|
C1ql4
|
complement C1q like 4 |
| chr1_-_100980340 | 0.12 |
ENSRNOT00000064272
|
Prmt1
|
protein arginine methyltransferase 1 |
| chr15_-_44860604 | 0.12 |
ENSRNOT00000018637
|
Nefm
|
neurofilament medium |
| chr15_+_2631529 | 0.12 |
ENSRNOT00000018897
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr10_+_10889488 | 0.12 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chrX_-_135041027 | 0.12 |
ENSRNOT00000006484
|
Zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr12_-_16934706 | 0.12 |
ENSRNOT00000001720
|
Mafk
|
MAF bZIP transcription factor K |
| chr10_+_89376530 | 0.12 |
ENSRNOT00000028089
|
Rnd2
|
Rho family GTPase 2 |
| chr10_-_56531483 | 0.12 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr12_+_8976585 | 0.12 |
ENSRNOT00000071796
|
LOC100911238
|
proteasome maturation protein-like |
| chr10_+_88459490 | 0.12 |
ENSRNOT00000080231
ENSRNOT00000067559 |
Ttc25
|
tetratricopeptide repeat domain 25 |
| chr14_+_34354503 | 0.12 |
ENSRNOT00000002941
|
Nmu
|
neuromedin U |
| chr2_-_235257464 | 0.11 |
ENSRNOT00000093472
|
Gar1
|
GAR1 ribonucleoprotein |
| chr1_-_84008293 | 0.11 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr2_+_197682000 | 0.11 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr7_-_13751271 | 0.11 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chr1_-_222178725 | 0.11 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr14_+_60764409 | 0.11 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr1_+_126979575 | 0.11 |
ENSRNOT00000016916
ENSRNOT00000081746 |
Selenos
|
selenoprotein S |
| chr4_-_117296082 | 0.11 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr19_+_55917736 | 0.11 |
ENSRNOT00000020635
|
Rpl13
|
ribosomal protein L13 |
| chr6_-_93562314 | 0.11 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr10_+_85257876 | 0.11 |
ENSRNOT00000014752
|
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr8_+_66296883 | 0.11 |
ENSRNOT00000018467
|
Tle3
|
transducin-like enhancer of split 3 |
| chr2_+_188561429 | 0.11 |
ENSRNOT00000076458
ENSRNOT00000027867 |
Krtcap2
|
keratinocyte associated protein 2 |
| chr10_+_64930023 | 0.11 |
ENSRNOT00000071102
|
AABR07030053.1
|
|
| chr5_-_138470096 | 0.11 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr10_-_13536899 | 0.11 |
ENSRNOT00000008537
|
Amdhd2
|
amidohydrolase domain containing 2 |
| chr17_-_66397653 | 0.11 |
ENSRNOT00000024098
|
Actn2
|
actinin alpha 2 |
| chr5_-_24631679 | 0.11 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr12_-_30268990 | 0.11 |
ENSRNOT00000079546
|
LOC103693015
|
vitamin K epoxide reductase complex subunit 1-like protein 1 |
| chr15_+_27850151 | 0.11 |
ENSRNOT00000087958
|
Apex1
|
apurinic/apyrimidinic endodeoxyribonuclease 1 |
| chr2_-_211017778 | 0.11 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr1_+_40879747 | 0.11 |
ENSRNOT00000083702
|
Akap12
|
A-kinase anchoring protein 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hey1_Myc_Mxi1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.2 | 0.8 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 0.5 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.4 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 3.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.6 | GO:2000851 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.3 | GO:1903544 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.1 | 0.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.3 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.1 | 0.2 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.1 | 0.2 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.1 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.2 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.2 | GO:2000771 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.1 | 0.2 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.3 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.2 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0034395 | heme oxidation(GO:0006788) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.2 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.2 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.3 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:2001137 | actin filament uncapping(GO:0051695) positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0060468 | peptidyl-tyrosine sulfation(GO:0006478) negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0072143 | renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.2 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of phagocytosis, engulfment(GO:0060101) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.0 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0061198 | maintenance of chromatin silencing(GO:0006344) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:1903189 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) positive regulation of superoxide dismutase activity(GO:1901671) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of removal of superoxide radicals(GO:1904833) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0034443 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.0 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.3 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.2 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.0 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0060683 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling(GO:0060683) embryonic lung development(GO:1990401) |
| 0.0 | 0.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.0 | 0.0 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.0 | GO:1904373 | response to kainic acid(GO:1904373) |
| 0.0 | 0.0 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0060631 | meiotic DNA double-strand break formation(GO:0042138) regulation of meiosis I(GO:0060631) |
| 0.0 | 0.0 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0046016 | positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.1 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0015746 | citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.1 | GO:1904874 | regulation of telomerase RNA localization to Cajal body(GO:1904872) positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.0 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.0 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:1904586 | response to sodium phosphate(GO:1904383) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.0 | GO:0046864 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.0 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0046671 | negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0044393 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 0.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.4 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 3.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0017098 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.0 | GO:0016823 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |