Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
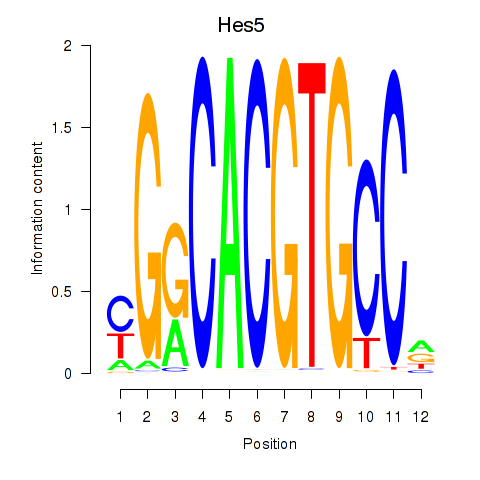
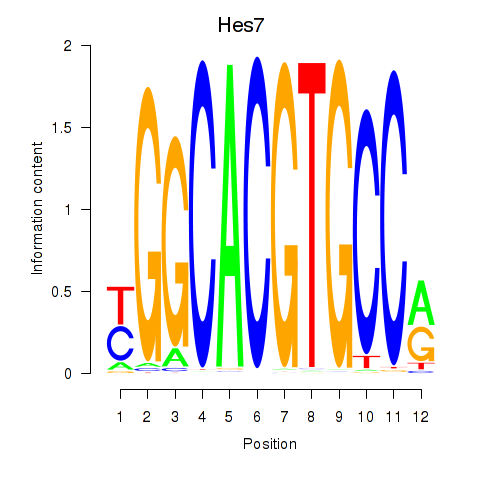
Results for Hes5_Hes7
Z-value: 0.41
Transcription factors associated with Hes5_Hes7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes5
|
ENSRNOG00000013850 | hes family bHLH transcription factor 5 |
|
Hes7
|
ENSRNOG00000007391 | hes family bHLH transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes7 | rn6_v1_chr10_+_55707164_55707164 | 0.42 | 4.8e-01 | Click! |
| Hes5 | rn6_v1_chr5_+_172364421_172364421 | -0.11 | 8.6e-01 | Click! |
Activity profile of Hes5_Hes7 motif
Sorted Z-values of Hes5_Hes7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_90315492 | 0.31 |
ENSRNOT00000070807
|
Gng4
|
G protein subunit gamma 4 |
| chr17_-_13393243 | 0.29 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr3_+_159569363 | 0.18 |
ENSRNOT00000064159
|
Tox2
|
TOX high mobility group box family member 2 |
| chr10_+_91710495 | 0.10 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr2_+_206064394 | 0.10 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr5_+_152681101 | 0.10 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr3_+_59981959 | 0.10 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr11_-_60613718 | 0.08 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr9_-_10054359 | 0.08 |
ENSRNOT00000072001
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr2_+_197682000 | 0.07 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr10_+_103438303 | 0.07 |
ENSRNOT00000085262
|
Cd300a
|
Cd300a molecule |
| chr10_+_83476107 | 0.07 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr5_+_64789456 | 0.06 |
ENSRNOT00000009584
|
Zfp189
|
zinc finger protein 189 |
| chr10_+_40208223 | 0.06 |
ENSRNOT00000000772
|
Hint1
|
histidine triad nucleotide binding protein 1 |
| chr2_+_147006830 | 0.06 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chr10_-_40375605 | 0.06 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr5_-_173098816 | 0.05 |
ENSRNOT00000023696
|
Mib2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr2_+_150211898 | 0.05 |
ENSRNOT00000018767
|
Sucnr1
|
succinate receptor 1 |
| chr11_+_47090245 | 0.05 |
ENSRNOT00000060645
|
AABR07033987.1
|
|
| chr6_-_59950586 | 0.05 |
ENSRNOT00000005800
|
Arl4a
|
ADP-ribosylation factor like GTPase 4A |
| chr10_+_10889488 | 0.05 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr3_+_138770574 | 0.05 |
ENSRNOT00000012510
ENSRNOT00000066986 |
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr5_-_156141537 | 0.05 |
ENSRNOT00000019004
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr1_-_239378095 | 0.05 |
ENSRNOT00000078466
|
AABR07006544.1
|
|
| chr12_-_16934706 | 0.05 |
ENSRNOT00000001720
|
Mafk
|
MAF bZIP transcription factor K |
| chr12_+_8976585 | 0.04 |
ENSRNOT00000071796
|
LOC100911238
|
proteasome maturation protein-like |
| chr12_-_8759599 | 0.04 |
ENSRNOT00000073155
|
Pomp
|
proteasome maturation protein |
| chr3_+_112346627 | 0.04 |
ENSRNOT00000074392
|
Snap23
|
synaptosomal-associated protein 23 |
| chr6_-_26486695 | 0.04 |
ENSRNOT00000073236
|
Krtcap3
|
keratinocyte associated protein 3 |
| chr20_-_5754773 | 0.04 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr3_-_92242318 | 0.04 |
ENSRNOT00000007018
|
Trim44
|
tripartite motif-containing 44 |
| chr20_+_4576514 | 0.04 |
ENSRNOT00000090125
ENSRNOT00000047370 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr9_+_20213776 | 0.04 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr5_-_156105867 | 0.04 |
ENSRNOT00000078851
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr2_-_93641497 | 0.03 |
ENSRNOT00000013720
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr1_-_220787238 | 0.03 |
ENSRNOT00000083656
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr3_-_81282157 | 0.03 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr18_+_62174670 | 0.03 |
ENSRNOT00000025362
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr8_-_59607275 | 0.03 |
ENSRNOT00000019307
|
Chrna3
|
cholinergic receptor nicotinic alpha 3 subunit |
| chr17_-_56909992 | 0.03 |
ENSRNOT00000021801
|
Bambi
|
BMP and activin membrane-bound inhibitor |
| chr2_-_196415530 | 0.03 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr1_-_164143818 | 0.03 |
ENSRNOT00000022557
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr19_+_57484634 | 0.03 |
ENSRNOT00000025695
|
Arv1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr4_-_157331905 | 0.03 |
ENSRNOT00000020647
|
Tpi1
|
triosephosphate isomerase 1 |
| chr10_-_20818128 | 0.03 |
ENSRNOT00000011061
|
Wwc1
|
WW and C2 domain containing 1 |
| chr5_+_157434481 | 0.03 |
ENSRNOT00000088556
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr9_+_80118029 | 0.03 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr2_-_93985378 | 0.03 |
ENSRNOT00000075493
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr7_-_12779862 | 0.03 |
ENSRNOT00000080292
|
Tmem259
|
transmembrane protein 259 |
| chr3_-_92290919 | 0.02 |
ENSRNOT00000007077
|
Fjx1
|
four jointed box 1 |
| chr9_-_15410943 | 0.02 |
ENSRNOT00000074217
|
Ccnd3
|
cyclin D3 |
| chr5_+_60809762 | 0.02 |
ENSRNOT00000016839
|
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr1_+_78546012 | 0.02 |
ENSRNOT00000058172
|
Gng5
|
G protein subunit gamma 5 |
| chr8_+_21669236 | 0.02 |
ENSRNOT00000027818
|
Pin1
|
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
| chr16_-_75855745 | 0.02 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr9_-_82419288 | 0.02 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr5_+_136112417 | 0.02 |
ENSRNOT00000025990
|
Tmem53
|
transmembrane protein 53 |
| chr4_+_157453069 | 0.02 |
ENSRNOT00000088622
|
Mlf2
|
myeloid leukemia factor 2 |
| chr1_+_82089922 | 0.02 |
ENSRNOT00000071251
|
Zfp526
|
zinc finger protein 526 |
| chr2_-_183031214 | 0.02 |
ENSRNOT00000013260
|
LOC679811
|
similar to RIKEN cDNA D930015E06 |
| chr20_+_12225202 | 0.02 |
ENSRNOT00000045315
|
Col18a1
|
collagen type XVIII alpha 1 chain |
| chr16_+_71889235 | 0.02 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr17_+_9837402 | 0.02 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr19_-_57422093 | 0.02 |
ENSRNOT00000025512
|
RGD1559896
|
similar to RIKEN cDNA 2310022B05 |
| chr4_+_157452607 | 0.02 |
ENSRNOT00000022467
|
Mlf2
|
myeloid leukemia factor 2 |
| chr10_-_36716601 | 0.02 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr20_+_4576057 | 0.02 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr4_-_179339795 | 0.02 |
ENSRNOT00000080410
|
Bcat1
|
branched chain amino acid transaminase 1 |
| chr1_+_224933920 | 0.02 |
ENSRNOT00000066823
|
Wdr74
|
WD repeat domain 74 |
| chr3_+_113415774 | 0.02 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr3_+_2288667 | 0.02 |
ENSRNOT00000012289
|
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr10_+_11338306 | 0.02 |
ENSRNOT00000033695
|
LOC100910875
|
protein FAM100A-like |
| chr5_+_141560192 | 0.02 |
ENSRNOT00000023354
|
Mycbp
|
Myc binding protein |
| chr2_+_211176556 | 0.02 |
ENSRNOT00000055880
|
Psrc1
|
proline and serine rich coiled-coil 1 |
| chr19_+_54314865 | 0.02 |
ENSRNOT00000024069
|
Irf8
|
interferon regulatory factor 8 |
| chr2_+_95320283 | 0.02 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr14_-_6786144 | 0.02 |
ENSRNOT00000091558
|
Mepe
|
matrix extracellular phosphoglycoprotein |
| chr5_+_165405168 | 0.01 |
ENSRNOT00000014934
|
Srm
|
spermidine synthase |
| chr7_-_12905339 | 0.01 |
ENSRNOT00000088330
|
Cdc34
|
cell division cycle 34 |
| chr5_-_36683356 | 0.01 |
ENSRNOT00000009043
|
Pou3f2
|
POU class 3 homeobox 2 |
| chr20_+_44436403 | 0.01 |
ENSRNOT00000000733
ENSRNOT00000076859 |
Fyn
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr7_+_99609191 | 0.01 |
ENSRNOT00000012883
|
Sqle
|
squalene epoxidase |
| chr7_-_130347587 | 0.01 |
ENSRNOT00000040541
|
Tymp
|
thymidine phosphorylase |
| chr4_-_157486844 | 0.01 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr5_-_59063592 | 0.01 |
ENSRNOT00000022400
|
Tln1
|
talin 1 |
| chr6_+_136050422 | 0.01 |
ENSRNOT00000080304
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr1_+_219390523 | 0.01 |
ENSRNOT00000054852
|
Gpr152
|
G protein-coupled receptor 152 |
| chr7_+_77899322 | 0.01 |
ENSRNOT00000006195
|
Fzd6
|
frizzled class receptor 6 |
| chr10_+_35133252 | 0.01 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr7_+_126619196 | 0.01 |
ENSRNOT00000030082
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr5_-_136112344 | 0.01 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr16_-_21347968 | 0.01 |
ENSRNOT00000068554
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr6_-_49851677 | 0.01 |
ENSRNOT00000007287
ENSRNOT00000077442 |
Acp1
|
acid phosphatase 1, soluble |
| chr19_-_25134055 | 0.01 |
ENSRNOT00000007519
|
Misp3
|
MISP family member 3 |
| chr14_+_84211600 | 0.01 |
ENSRNOT00000005996
|
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr1_+_102017263 | 0.01 |
ENSRNOT00000028680
|
Nomo1
|
nodal modulator 1 |
| chr1_+_247871807 | 0.01 |
ENSRNOT00000089563
|
Mlana
|
melan-A |
| chr2_+_189400696 | 0.01 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr4_-_157679962 | 0.01 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr11_+_64861768 | 0.01 |
ENSRNOT00000038439
|
Adprh
|
ADP-ribosylarginine hydrolase |
| chr13_+_71179910 | 0.01 |
ENSRNOT00000032236
|
Rgs16
|
regulator of G-protein signaling 16 |
| chr11_+_24266345 | 0.01 |
ENSRNOT00000046973
|
Jam2
|
junctional adhesion molecule 2 |
| chr11_+_60073383 | 0.01 |
ENSRNOT00000087508
|
Tagln3
|
transgelin 3 |
| chr20_-_48758905 | 0.01 |
ENSRNOT00000000710
|
Ddo
|
D-aspartate oxidase |
| chr4_-_153465203 | 0.01 |
ENSRNOT00000016776
|
Bid
|
BH3 interacting domain death agonist |
| chr19_-_46167950 | 0.01 |
ENSRNOT00000071024
|
AABR07043892.1
|
|
| chr2_+_85377318 | 0.01 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr20_-_43108198 | 0.01 |
ENSRNOT00000000742
|
Hdac2
|
histone deacetylase 2 |
| chr3_+_80021440 | 0.01 |
ENSRNOT00000018620
|
Acp2
|
acid phosphatase 2, lysosomal |
| chr19_-_57484742 | 0.00 |
ENSRNOT00000079783
|
Ttc13
|
tetratricopeptide repeat domain 13 |
| chr12_-_41485122 | 0.00 |
ENSRNOT00000001859
|
Ddx54
|
DEAD-box helicase 54 |
| chr6_-_36940868 | 0.00 |
ENSRNOT00000006470
|
Gen1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr18_+_72005581 | 0.00 |
ENSRNOT00000072519
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr7_+_77678968 | 0.00 |
ENSRNOT00000006917
|
Atp6v1c1
|
ATPase H+ transporting V1 subunit C1 |
| chr10_-_84789832 | 0.00 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr20_+_11168298 | 0.00 |
ENSRNOT00000032240
|
Trappc10
|
trafficking protein particle complex 10 |
| chr14_+_84465515 | 0.00 |
ENSRNOT00000035591
|
Osm
|
oncostatin M |
| chr7_+_77966722 | 0.00 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr16_-_19349080 | 0.00 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr7_+_126096793 | 0.00 |
ENSRNOT00000047234
ENSRNOT00000088042 |
Fbln1
|
fibulin 1 |
| chr8_+_128972311 | 0.00 |
ENSRNOT00000025460
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr19_-_57484566 | 0.00 |
ENSRNOT00000025548
|
Ttc13
|
tetratricopeptide repeat domain 13 |
| chr20_-_13458110 | 0.00 |
ENSRNOT00000072183
|
Slc5a4
|
solute carrier family 5 member 4 |
| chr1_+_185673177 | 0.00 |
ENSRNOT00000048020
|
Sox6
|
SRY box 6 |
| chr7_-_2986935 | 0.00 |
ENSRNOT00000081125
ENSRNOT00000006578 |
Pa2g4
|
proliferation-associated 2G4 |
| chr19_-_25914689 | 0.00 |
ENSRNOT00000091994
ENSRNOT00000039354 |
Nf1x
|
nuclear factor 1 X |
| chr12_+_37805332 | 0.00 |
ENSRNOT00000048528
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr1_-_282251257 | 0.00 |
ENSRNOT00000015186
|
Prdx3
|
peroxiredoxin 3 |
| chr10_-_15960673 | 0.00 |
ENSRNOT00000036447
|
RGD1311343
|
similar to RIKEN cDNA 4930524B15 |
| chr3_+_138508899 | 0.00 |
ENSRNOT00000009437
|
Kat14
|
lysine acetyltransferase 14 |
| chr7_+_60316045 | 0.00 |
ENSRNOT00000064594
|
Lyc2
|
lysozyme C type 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes5_Hes7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of phagocytosis, engulfment(GO:0060101) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) phenotypic switching(GO:0036166) |
| 0.0 | 0.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0060631 | meiotic DNA double-strand break formation(GO:0042138) regulation of meiosis I(GO:0060631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |