Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
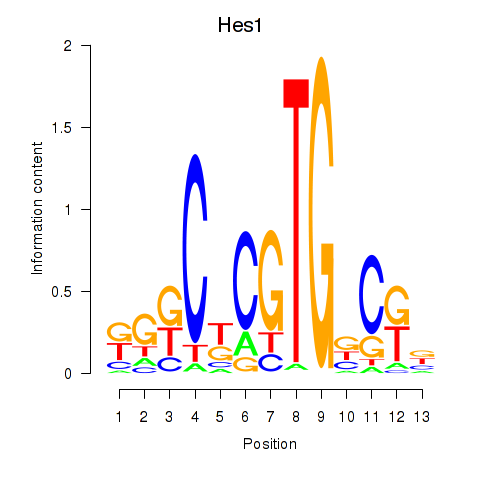
Results for Hes1
Z-value: 0.67
Transcription factors associated with Hes1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes1
|
ENSRNOG00000001720 | hes family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes1 | rn6_v1_chr11_-_74315248_74315248 | 0.69 | 2.0e-01 | Click! |
Activity profile of Hes1 motif
Sorted Z-values of Hes1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_156811650 | 0.77 |
ENSRNOT00000068065
|
Fam43b
|
family with sequence similarity 43, member B |
| chr4_-_117296082 | 0.50 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr3_+_147585947 | 0.44 |
ENSRNOT00000006833
|
Scrt2
|
scratch family transcriptional repressor 2 |
| chr5_+_156810991 | 0.23 |
ENSRNOT00000055904
|
AABR07050222.1
|
|
| chr6_-_42473738 | 0.20 |
ENSRNOT00000033327
|
Kcnf1
|
potassium voltage-gated channel modifier subfamily F member 1 |
| chr15_+_51756978 | 0.19 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr17_-_23923792 | 0.19 |
ENSRNOT00000018739
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr4_+_32373641 | 0.17 |
ENSRNOT00000076086
|
Dlx6
|
distal-less homeobox 6 |
| chr3_+_154395187 | 0.16 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr8_-_115981910 | 0.16 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr7_+_12764993 | 0.16 |
ENSRNOT00000017064
|
Grin3b
|
glutamate ionotropic receptor NMDA type subunit 3B |
| chr7_-_70452675 | 0.16 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr2_+_124400679 | 0.16 |
ENSRNOT00000058495
ENSRNOT00000067034 |
Spry1
|
sprouty RTK signaling antagonist 1 |
| chr11_-_36479868 | 0.15 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr4_+_148139528 | 0.15 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr6_+_136720582 | 0.15 |
ENSRNOT00000086868
|
Kif26a
|
kinesin family member 26A |
| chr15_-_61564695 | 0.15 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr10_-_29450644 | 0.14 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr10_+_17209212 | 0.14 |
ENSRNOT00000073675
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr11_-_38420032 | 0.14 |
ENSRNOT00000002209
ENSRNOT00000080681 |
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr3_+_13838304 | 0.14 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr19_+_10519493 | 0.13 |
ENSRNOT00000030229
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr18_-_56115593 | 0.13 |
ENSRNOT00000045041
|
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr7_+_11215328 | 0.13 |
ENSRNOT00000061194
|
LOC690617
|
hypothetical protein LOC690617 |
| chr15_-_34198921 | 0.13 |
ENSRNOT00000024991
|
Nrl
|
neural retina leucine zipper |
| chr3_-_38277440 | 0.12 |
ENSRNOT00000037857
|
Stam2
|
signal transducing adaptor molecule 2 |
| chr15_+_24078280 | 0.12 |
ENSRNOT00000015511
ENSRNOT00000063807 |
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr6_+_136720266 | 0.12 |
ENSRNOT00000018278
|
Kif26a
|
kinesin family member 26A |
| chr8_+_118333706 | 0.12 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr7_+_130474279 | 0.12 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr5_+_48290061 | 0.12 |
ENSRNOT00000076508
|
Ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr12_-_41485122 | 0.11 |
ENSRNOT00000001859
|
Ddx54
|
DEAD-box helicase 54 |
| chr8_+_70603249 | 0.11 |
ENSRNOT00000067016
ENSRNOT00000072486 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr2_+_95320283 | 0.11 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr5_-_152589719 | 0.11 |
ENSRNOT00000022522
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr4_+_99239115 | 0.11 |
ENSRNOT00000009515
|
Cd8a
|
CD8a molecule |
| chr10_-_108196217 | 0.10 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr10_-_109821807 | 0.10 |
ENSRNOT00000054949
|
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr2_-_211715311 | 0.10 |
ENSRNOT00000027738
|
Prpf38b
|
pre-mRNA processing factor 38B |
| chr1_+_222519615 | 0.10 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr15_+_27875911 | 0.10 |
ENSRNOT00000013582
|
Pnp
|
purine nucleoside phosphorylase |
| chr4_-_132171153 | 0.10 |
ENSRNOT00000015058
ENSRNOT00000015075 |
Prok2
|
prokineticin 2 |
| chr3_+_138504214 | 0.09 |
ENSRNOT00000091529
|
Kat14
|
lysine acetyltransferase 14 |
| chr7_-_11330278 | 0.09 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr17_-_20364714 | 0.08 |
ENSRNOT00000070962
|
Jarid2
|
jumonji and AT-rich interaction domain containing 2 |
| chr17_+_76079720 | 0.08 |
ENSRNOT00000073933
|
Proser2
|
proline and serine rich 2 |
| chr1_-_125967756 | 0.08 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr2_-_235257464 | 0.08 |
ENSRNOT00000093472
|
Gar1
|
GAR1 ribonucleoprotein |
| chr11_+_39482408 | 0.08 |
ENSRNOT00000075126
|
Hmgn1
|
high mobility group nucleosome binding domain 1 |
| chr14_-_78377825 | 0.08 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr17_-_85141210 | 0.08 |
ENSRNOT00000000162
|
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr1_+_248228496 | 0.08 |
ENSRNOT00000015406
|
Uhrf2
|
ubiquitin like with PHD and ring finger domains 2 |
| chr7_-_126461658 | 0.07 |
ENSRNOT00000081032
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr1_-_212633304 | 0.07 |
ENSRNOT00000025611
|
Olr286
|
olfactory receptor 286 |
| chr1_+_82452469 | 0.07 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr10_+_15620871 | 0.07 |
ENSRNOT00000089435
ENSRNOT00000066430 |
Nprl3
|
NPR3-like, GATOR1 complex subunit |
| chr13_+_52662996 | 0.07 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr10_+_48773828 | 0.07 |
ENSRNOT00000004113
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr20_-_31072469 | 0.07 |
ENSRNOT00000082448
|
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr9_+_10013854 | 0.07 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr13_-_102942863 | 0.07 |
ENSRNOT00000003198
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr1_-_86948845 | 0.06 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr5_+_152681101 | 0.06 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr8_+_82288705 | 0.06 |
ENSRNOT00000012409
|
Bcl2l10
|
BCL2 like 10 |
| chr1_+_162320730 | 0.06 |
ENSRNOT00000035743
|
Kctd21
|
potassium channel tetramerization domain containing 21 |
| chr15_+_19338175 | 0.06 |
ENSRNOT00000075209
|
Ptger2
|
prostaglandin E receptor 2 |
| chr20_+_10930518 | 0.06 |
ENSRNOT00000001589
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr15_+_19603288 | 0.06 |
ENSRNOT00000035491
|
LOC102552640
|
REST corepressor 2-like |
| chr10_-_72142533 | 0.06 |
ENSRNOT00000030885
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr12_+_47218969 | 0.06 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr3_+_150910398 | 0.06 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr8_+_62723788 | 0.06 |
ENSRNOT00000010620
|
Sema7a
|
semaphorin 7A (John Milton Hagen blood group) |
| chr2_-_210550490 | 0.06 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr10_+_47018974 | 0.06 |
ENSRNOT00000079375
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr10_+_103438303 | 0.06 |
ENSRNOT00000085262
|
Cd300a
|
Cd300a molecule |
| chr11_+_34051993 | 0.05 |
ENSRNOT00000076473
ENSRNOT00000064751 |
Morc3
|
MORC family CW-type zinc finger 3 |
| chr1_+_82480195 | 0.05 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr3_-_94419048 | 0.05 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr5_-_151117042 | 0.05 |
ENSRNOT00000066549
|
Fam76a
|
family with sequence similarity 76, member A |
| chr5_+_135735825 | 0.05 |
ENSRNOT00000068267
|
Zswim5
|
zinc finger, SWIM-type containing 5 |
| chr16_+_9486832 | 0.05 |
ENSRNOT00000082208
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr10_+_102136283 | 0.05 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr9_+_94310469 | 0.05 |
ENSRNOT00000065743
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr10_-_15161938 | 0.05 |
ENSRNOT00000026641
|
Fam173a
|
family with sequence similarity 173, member A |
| chr4_+_116968000 | 0.05 |
ENSRNOT00000020786
|
Emx1
|
empty spiracles homeobox 1 |
| chr6_-_26855658 | 0.05 |
ENSRNOT00000011635
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr9_+_10428853 | 0.05 |
ENSRNOT00000074253
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr12_-_13998172 | 0.05 |
ENSRNOT00000001476
|
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr3_-_4405194 | 0.05 |
ENSRNOT00000072025
|
AABR07051266.1
|
|
| chr14_-_85350786 | 0.05 |
ENSRNOT00000012634
|
LOC100912481
|
RNA-binding protein EWS-like |
| chr1_+_198911911 | 0.04 |
ENSRNOT00000055002
|
Prr14
|
proline rich 14 |
| chr8_+_59278262 | 0.04 |
ENSRNOT00000017053
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr5_+_154205402 | 0.04 |
ENSRNOT00000064925
|
Srsf10
|
serine and arginine rich splicing factor 10 |
| chr1_-_18058055 | 0.04 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr5_-_154363329 | 0.04 |
ENSRNOT00000064596
|
Eloa
|
elongin A |
| chrX_+_128416722 | 0.04 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr3_-_175479395 | 0.04 |
ENSRNOT00000077308
|
Hrh3
|
histamine receptor H3 |
| chr10_+_14547172 | 0.04 |
ENSRNOT00000092043
|
Unkl
|
unkempt family like zinc finger |
| chr7_-_2986935 | 0.04 |
ENSRNOT00000081125
ENSRNOT00000006578 |
Pa2g4
|
proliferation-associated 2G4 |
| chr2_-_189400323 | 0.04 |
ENSRNOT00000024364
|
Ubap2l
|
ubiquitin associated protein 2-like |
| chr17_-_89923990 | 0.04 |
ENSRNOT00000071150
|
LOC100910957
|
acyl-CoA-binding domain-containing protein 5-like |
| chr1_+_128199322 | 0.04 |
ENSRNOT00000075131
|
Lysmd4
|
LysM domain containing 4 |
| chr5_+_150080072 | 0.04 |
ENSRNOT00000047917
|
Tmem200b
|
transmembrane protein 200B |
| chr6_-_28464118 | 0.04 |
ENSRNOT00000068214
|
Efr3b
|
EFR3 homolog B |
| chr17_-_89923423 | 0.04 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr12_-_39396042 | 0.04 |
ENSRNOT00000001746
|
P2rx7
|
purinergic receptor P2X 7 |
| chr2_-_22385855 | 0.04 |
ENSRNOT00000083531
|
Thbs4
|
thrombospondin 4 |
| chr6_+_93539271 | 0.04 |
ENSRNOT00000078791
|
Tomm20l
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr16_-_71237118 | 0.04 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr10_+_63803309 | 0.03 |
ENSRNOT00000036666
|
Myo1c
|
myosin 1C |
| chrX_+_156716604 | 0.03 |
ENSRNOT00000092207
|
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr3_+_111013856 | 0.03 |
ENSRNOT00000016854
|
Zfyve19
|
zinc finger FYVE-type containing 19 |
| chr5_-_163167299 | 0.03 |
ENSRNOT00000022478
|
Tnfrsf1b
|
TNF receptor superfamily member 1B |
| chr20_-_5533600 | 0.03 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr2_-_235257731 | 0.03 |
ENSRNOT00000078074
|
Gar1
|
GAR1 ribonucleoprotein |
| chr14_-_85088523 | 0.03 |
ENSRNOT00000010690
|
Nf2
|
neurofibromin 2 |
| chr20_-_5533448 | 0.03 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr1_-_167308827 | 0.03 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr1_+_214534284 | 0.03 |
ENSRNOT00000064254
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr3_+_160945359 | 0.03 |
ENSRNOT00000019761
|
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr9_+_50247692 | 0.03 |
ENSRNOT00000023029
|
Nck2
|
NCK adaptor protein 2 |
| chr5_-_144221263 | 0.03 |
ENSRNOT00000013570
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr13_-_88061108 | 0.03 |
ENSRNOT00000003774
|
Rgs4
|
regulator of G-protein signaling 4 |
| chr20_+_11168298 | 0.03 |
ENSRNOT00000032240
|
Trappc10
|
trafficking protein particle complex 10 |
| chr2_-_250232295 | 0.03 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr7_+_125833557 | 0.03 |
ENSRNOT00000018155
|
Nup50
|
nucleoporin 50 |
| chr1_-_72727112 | 0.03 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr17_-_84830185 | 0.03 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chr16_+_48120864 | 0.03 |
ENSRNOT00000068562
|
Stox2
|
storkhead box 2 |
| chr16_+_59564419 | 0.03 |
ENSRNOT00000083434
|
Lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr9_-_114709546 | 0.03 |
ENSRNOT00000026177
|
Rab12
|
RAB12, member RAS oncogene family |
| chr10_+_88459490 | 0.03 |
ENSRNOT00000080231
ENSRNOT00000067559 |
Ttc25
|
tetratricopeptide repeat domain 25 |
| chr16_+_71787966 | 0.03 |
ENSRNOT00000080084
|
Htra4
|
HtrA serine peptidase 4 |
| chr5_-_154319629 | 0.03 |
ENSRNOT00000014415
|
Lypla2
|
lysophospholipase II |
| chr4_+_168689163 | 0.03 |
ENSRNOT00000049848
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr8_+_65733400 | 0.03 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr3_-_176791960 | 0.03 |
ENSRNOT00000018237
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr2_-_22385676 | 0.03 |
ENSRNOT00000065224
|
Thbs4
|
thrombospondin 4 |
| chr7_+_70452579 | 0.03 |
ENSRNOT00000046099
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr3_-_22952063 | 0.03 |
ENSRNOT00000083850
|
Psmb7
|
proteasome subunit beta 7 |
| chr8_-_122402127 | 0.02 |
ENSRNOT00000074238
ENSRNOT00000076548 |
Crtapl1
|
cartilage associated protein-like 1 |
| chr8_-_111850393 | 0.02 |
ENSRNOT00000044956
|
Cdv3
|
CDV3 homolog |
| chr10_-_105600913 | 0.02 |
ENSRNOT00000068026
|
Rhbdf2
|
rhomboid 5 homolog 2 |
| chr10_+_57462447 | 0.02 |
ENSRNOT00000041443
|
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr2_+_113984646 | 0.02 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr10_-_4910305 | 0.02 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr11_-_60613718 | 0.02 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr9_-_42839837 | 0.02 |
ENSRNOT00000038610
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr7_+_126939925 | 0.02 |
ENSRNOT00000022746
|
Gramd4
|
GRAM domain containing 4 |
| chr10_+_108750620 | 0.02 |
ENSRNOT00000005337
|
Rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr1_-_143535583 | 0.02 |
ENSRNOT00000087785
|
Homer2
|
homer scaffolding protein 2 |
| chr4_-_66002444 | 0.02 |
ENSRNOT00000018645
|
Zc3hav1l
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr6_-_26771164 | 0.02 |
ENSRNOT00000009090
|
Tcf23
|
transcription factor 23 |
| chr14_+_44413636 | 0.02 |
ENSRNOT00000003552
|
Smim14
|
small integral membrane protein 14 |
| chr14_+_113968563 | 0.02 |
ENSRNOT00000006085
|
Clhc1
|
clathrin heavy chain linker domain containing 1 |
| chrX_+_63343546 | 0.02 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr7_+_77899322 | 0.02 |
ENSRNOT00000006195
|
Fzd6
|
frizzled class receptor 6 |
| chr10_-_76263866 | 0.02 |
ENSRNOT00000003219
|
Scpep1
|
serine carboxypeptidase 1 |
| chr4_+_84854386 | 0.02 |
ENSRNOT00000013620
|
Mturn
|
maturin, neural progenitor differentiation regulator homolog |
| chr19_-_39646693 | 0.02 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr7_+_12542713 | 0.02 |
ENSRNOT00000080089
|
Med16
|
mediator complex subunit 16 |
| chr10_-_109757550 | 0.02 |
ENSRNOT00000054957
|
Arhgdia
|
Rho GDP dissociation inhibitor alpha |
| chr16_+_20668971 | 0.02 |
ENSRNOT00000027073
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr7_+_66595742 | 0.02 |
ENSRNOT00000031191
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr19_+_17290178 | 0.01 |
ENSRNOT00000060865
|
Aktip
|
AKT interacting protein |
| chr7_-_119689938 | 0.01 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr5_-_39215102 | 0.01 |
ENSRNOT00000050653
|
Klhl32
|
kelch-like family member 32 |
| chr20_+_2501252 | 0.01 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr1_-_90000386 | 0.01 |
ENSRNOT00000037519
|
Wtip
|
Wilms tumor 1 interacting protein |
| chr13_+_89797800 | 0.01 |
ENSRNOT00000005811
|
Usf1
|
upstream transcription factor 1 |
| chr1_-_257498844 | 0.01 |
ENSRNOT00000019056
|
Noc3l
|
NOC3-like DNA replication regulator |
| chr8_-_23482939 | 0.01 |
ENSRNOT00000065367
|
Rp9
|
retinitis pigmentosa 9 |
| chr15_-_52029816 | 0.01 |
ENSRNOT00000013067
|
Slc39a14
|
solute carrier family 39 member 14 |
| chr9_-_88357182 | 0.01 |
ENSRNOT00000041176
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr14_+_86101277 | 0.01 |
ENSRNOT00000018846
|
Aebp1
|
AE binding protein 1 |
| chr2_+_187893368 | 0.01 |
ENSRNOT00000092031
|
Mex3a
|
mex-3 RNA binding family member A |
| chr1_-_188373571 | 0.01 |
ENSRNOT00000075020
|
Knop1
|
lysine-rich nucleolar protein 1 |
| chr11_-_24641820 | 0.01 |
ENSRNOT00000044081
ENSRNOT00000048854 |
App
|
amyloid beta precursor protein |
| chr12_+_49626871 | 0.01 |
ENSRNOT00000082593
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr8_+_57964988 | 0.01 |
ENSRNOT00000009497
|
Kdelc2
|
KDEL motif containing 2 |
| chr17_+_70684340 | 0.01 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr19_+_11473541 | 0.01 |
ENSRNOT00000082066
|
Amfr
|
autocrine motility factor receptor |
| chr14_-_86297623 | 0.01 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chr7_+_78188912 | 0.01 |
ENSRNOT00000043079
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr8_-_112807598 | 0.01 |
ENSRNOT00000015344
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr12_+_25036605 | 0.01 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chr2_+_251634431 | 0.01 |
ENSRNOT00000045016
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_-_11144641 | 0.01 |
ENSRNOT00000081246
|
Slc12a3
|
solute carrier family 12 member 3 |
| chr10_+_104437648 | 0.01 |
ENSRNOT00000035001
|
AC130970.1
|
|
| chr4_+_119997268 | 0.01 |
ENSRNOT00000073429
|
Rpn1
|
ribophorin I |
| chr20_-_13706205 | 0.01 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr7_-_28040510 | 0.01 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr3_-_112371664 | 0.01 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chr16_+_20121352 | 0.01 |
ENSRNOT00000025347
|
Insl3
|
insulin-like 3 |
| chr5_+_63050758 | 0.01 |
ENSRNOT00000009452
|
Tgfbr1
|
transforming growth factor, beta receptor 1 |
| chr2_-_183487820 | 0.00 |
ENSRNOT00000066477
|
Fhdc1
|
FH2 domain containing 1 |
| chr2_-_179704629 | 0.00 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr2_+_9578345 | 0.00 |
ENSRNOT00000021756
|
Mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr2_+_207108552 | 0.00 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr13_+_44475970 | 0.00 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.2 | GO:0060938 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.2 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0060101 | negative regulation of phagocytosis, engulfment(GO:0060101) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.0 | GO:0043132 | phospholipid transfer to membrane(GO:0006649) NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) |
| 0.0 | 0.2 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0071051 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0004731 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |