Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
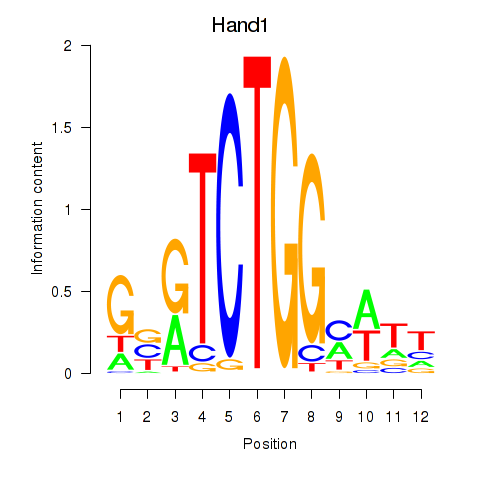
Results for Hand1
Z-value: 0.30
Transcription factors associated with Hand1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hand1
|
ENSRNOG00000002582 | heart and neural crest derivatives expressed 1 |
Activity profile of Hand1 motif
Sorted Z-values of Hand1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_5067330 | 0.27 |
ENSRNOT00000037482
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr4_+_86275717 | 0.15 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr7_+_123168811 | 0.15 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr1_+_198690794 | 0.15 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr7_+_142905758 | 0.12 |
ENSRNOT00000078663
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_261446570 | 0.12 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr16_+_7303578 | 0.11 |
ENSRNOT00000025656
|
Sema3g
|
semaphorin 3G |
| chr20_+_5094468 | 0.10 |
ENSRNOT00000078576
|
Abhd16a
|
abhydrolase domain containing 16A |
| chr1_+_86873253 | 0.10 |
ENSRNOT00000071239
|
Fbxo27
|
F-box protein 27 |
| chr1_-_216080287 | 0.09 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr10_-_64657089 | 0.09 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr16_-_73827488 | 0.08 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr11_+_83048636 | 0.08 |
ENSRNOT00000002408
|
RGD1562339
|
RGD1562339 |
| chr12_-_46889082 | 0.08 |
ENSRNOT00000001525
|
Pla2g1b
|
phospholipase A2 group IB |
| chr6_-_136379348 | 0.07 |
ENSRNOT00000016208
|
Xrcc3
|
X-ray repair cross complementing 3 |
| chr3_-_11410732 | 0.07 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr3_+_110982553 | 0.07 |
ENSRNOT00000030754
|
LOC691418
|
hypothetical protein LOC691418 |
| chr3_+_175144495 | 0.07 |
ENSRNOT00000082601
ENSRNOT00000088026 |
Cdh4
|
cadherin 4 |
| chr10_-_38838272 | 0.07 |
ENSRNOT00000089495
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr8_-_119012671 | 0.07 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr8_+_44847157 | 0.06 |
ENSRNOT00000080288
|
Clmp
|
CXADR-like membrane protein |
| chr3_+_72540538 | 0.06 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr6_-_15191660 | 0.06 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr1_+_218076116 | 0.06 |
ENSRNOT00000028374
|
Oraov1
|
oral cancer overexpressed 1 |
| chr15_-_26678420 | 0.06 |
ENSRNOT00000041420
|
RGD1310110
|
similar to 3632451O06Rik protein |
| chr5_+_152613255 | 0.06 |
ENSRNOT00000037097
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr8_+_70603249 | 0.06 |
ENSRNOT00000067016
ENSRNOT00000072486 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr11_+_72044096 | 0.05 |
ENSRNOT00000034757
|
Senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr3_+_101010899 | 0.05 |
ENSRNOT00000073258
|
Lin7c
|
lin-7 homolog C, crumbs cell polarity complex component |
| chr1_+_72661211 | 0.05 |
ENSRNOT00000033197
|
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr1_-_275906686 | 0.05 |
ENSRNOT00000092170
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr4_+_66276835 | 0.05 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr4_-_117575154 | 0.05 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr1_+_88182585 | 0.05 |
ENSRNOT00000044145
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr13_+_52624878 | 0.05 |
ENSRNOT00000076054
ENSRNOT00000076299 |
Tnni1
|
troponin I1, slow skeletal type |
| chr17_-_10818835 | 0.05 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr16_+_35573058 | 0.05 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr20_+_3246739 | 0.05 |
ENSRNOT00000061299
|
RT1-T24-2
|
RT1 class I, locus T24, gene 2 |
| chr13_-_91872954 | 0.05 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr1_+_44446765 | 0.05 |
ENSRNOT00000030132
|
Cldn20
|
claudin 20 |
| chr8_+_118662335 | 0.05 |
ENSRNOT00000029755
|
Ngp
|
neutrophilic granule protein |
| chr1_+_217173199 | 0.05 |
ENSRNOT00000075078
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr17_+_12762752 | 0.04 |
ENSRNOT00000044842
|
Diras2
|
DIRAS family GTPase 2 |
| chr15_-_48445588 | 0.04 |
ENSRNOT00000077197
ENSRNOT00000018583 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr18_+_74299931 | 0.04 |
ENSRNOT00000078403
|
Epg5
|
ectopic P-granules autophagy protein 5 homolog |
| chr14_+_60657686 | 0.04 |
ENSRNOT00000070892
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr14_+_2613406 | 0.04 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr3_-_3541947 | 0.04 |
ENSRNOT00000024786
|
Nacc2
|
NACC family member 2 |
| chr11_+_9642365 | 0.04 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr10_+_106264434 | 0.04 |
ENSRNOT00000003891
|
Sept9
|
septin 9 |
| chr12_+_16950728 | 0.04 |
ENSRNOT00000060370
|
Ints1
|
integrator complex subunit 1 |
| chr1_+_199196059 | 0.04 |
ENSRNOT00000090428
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr12_+_18074033 | 0.04 |
ENSRNOT00000001727
|
LOC103689975
|
integrator complex subunit 1-like |
| chr4_-_155959993 | 0.04 |
ENSRNOT00000029072
|
Clec4a1
|
C-type lectin domain family 4, member A1 |
| chr3_+_62648447 | 0.04 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chrX_-_118998300 | 0.04 |
ENSRNOT00000025575
|
AABR07041089.1
|
|
| chr1_-_88826302 | 0.04 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr20_+_4355175 | 0.04 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr16_+_50152008 | 0.04 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr8_+_130538651 | 0.04 |
ENSRNOT00000026343
|
Ackr2
|
atypical chemokine receptor 2 |
| chr10_-_95434688 | 0.04 |
ENSRNOT00000074809
|
AABR07030603.1
|
|
| chr9_+_92859988 | 0.03 |
ENSRNOT00000085332
|
Cab39
|
calcium binding protein 39 |
| chr2_+_198303168 | 0.03 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr17_+_28504623 | 0.03 |
ENSRNOT00000021568
|
F13a1
|
coagulation factor XIII A1 chain |
| chr2_+_144646308 | 0.03 |
ENSRNOT00000078337
ENSRNOT00000093407 |
Dclk1
|
doublecortin-like kinase 1 |
| chr17_+_23135985 | 0.03 |
ENSRNOT00000090794
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr7_+_120176530 | 0.03 |
ENSRNOT00000087548
|
Triobp
|
TRIO and F-actin binding protein |
| chr1_+_87180624 | 0.03 |
ENSRNOT00000056933
ENSRNOT00000027987 |
LOC103689986
|
protein YIF1B |
| chr20_-_2083688 | 0.03 |
ENSRNOT00000001011
|
Znrd1as1
|
ZNRD1 antisense RNA 1 |
| chr10_+_69434941 | 0.03 |
ENSRNOT00000009756
|
Ccl11
|
C-C motif chemokine ligand 11 |
| chr5_-_60559329 | 0.03 |
ENSRNOT00000017046
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr3_+_65816569 | 0.03 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr3_-_94182714 | 0.03 |
ENSRNOT00000015073
|
LOC100362814
|
hypothetical protein LOC100362814 |
| chr4_-_123510217 | 0.03 |
ENSRNOT00000080734
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr6_+_143938040 | 0.03 |
ENSRNOT00000005876
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr12_-_46809849 | 0.03 |
ENSRNOT00000081134
|
Pxn
|
paxillin |
| chr11_-_70961358 | 0.03 |
ENSRNOT00000002427
|
Lmln
|
leishmanolysin like peptidase |
| chr14_-_112946204 | 0.03 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr9_-_121931564 | 0.03 |
ENSRNOT00000056243
|
Tyms
|
thymidylate synthetase |
| chr10_-_90240509 | 0.03 |
ENSRNOT00000028407
|
Atxn7l3
|
ataxin 7-like 3 |
| chr5_-_158439078 | 0.03 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr6_+_12362813 | 0.03 |
ENSRNOT00000022370
|
Ston1
|
stonin 1 |
| chr10_+_34185898 | 0.03 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr12_+_15857805 | 0.03 |
ENSRNOT00000001667
|
Iqce
|
IQ motif containing E |
| chr1_+_27476375 | 0.03 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr8_+_58347736 | 0.03 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr16_+_20432899 | 0.03 |
ENSRNOT00000026271
|
Mpv17l2
|
MPV17 mitochondrial inner membrane protein like 2 |
| chr7_+_18409147 | 0.03 |
ENSRNOT00000086336
ENSRNOT00000011849 |
Adamts10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr12_+_49665282 | 0.03 |
ENSRNOT00000086397
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr2_+_248398917 | 0.03 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr2_+_58448917 | 0.03 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr2_+_200572502 | 0.03 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr8_-_132027006 | 0.03 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr3_+_162181974 | 0.03 |
ENSRNOT00000030203
|
Slc2a10
|
solute carrier family 2 member 10 |
| chr14_-_33677031 | 0.03 |
ENSRNOT00000002908
|
RGD1311575
|
hypothetical LOC289568 |
| chr1_+_88182155 | 0.03 |
ENSRNOT00000083176
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr8_-_50526843 | 0.03 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chrX_+_80213332 | 0.03 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr4_+_34282625 | 0.03 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chr10_+_105500290 | 0.03 |
ENSRNOT00000079080
ENSRNOT00000083593 |
Sphk1
|
sphingosine kinase 1 |
| chr1_-_101008266 | 0.03 |
ENSRNOT00000081430
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chr7_+_54247460 | 0.03 |
ENSRNOT00000005361
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr6_+_136720266 | 0.03 |
ENSRNOT00000018278
|
Kif26a
|
kinesin family member 26A |
| chr8_-_55467579 | 0.03 |
ENSRNOT00000040162
ENSRNOT00000088607 |
Layn
|
layilin |
| chr1_+_84067938 | 0.03 |
ENSRNOT00000057213
|
Numbl
|
NUMB-like, endocytic adaptor protein |
| chr14_+_108826831 | 0.03 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr13_-_52197205 | 0.03 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr7_+_144080614 | 0.03 |
ENSRNOT00000077687
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr12_-_37047628 | 0.03 |
ENSRNOT00000001337
|
Rflna
|
refilin A |
| chr6_-_21112734 | 0.02 |
ENSRNOT00000079819
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr6_+_44230985 | 0.02 |
ENSRNOT00000005858
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr12_-_50313976 | 0.02 |
ENSRNOT00000000824
|
Hps4
|
Hermansky-Pudlak syndrome 4 |
| chr1_+_225015616 | 0.02 |
ENSRNOT00000026416
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr15_+_43298794 | 0.02 |
ENSRNOT00000012736
|
Adra1a
|
adrenoceptor alpha 1A |
| chr7_-_118396728 | 0.02 |
ENSRNOT00000066431
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr11_+_61662270 | 0.02 |
ENSRNOT00000079521
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr1_-_90962466 | 0.02 |
ENSRNOT00000080613
|
LOC108348106
|
alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 |
| chr8_-_125645898 | 0.02 |
ENSRNOT00000036672
|
Rbms3
|
RNA binding motif, single stranded interacting protein 3 |
| chr16_+_70081035 | 0.02 |
ENSRNOT00000016098
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr1_-_125967756 | 0.02 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr12_+_37211316 | 0.02 |
ENSRNOT00000032739
|
Ccdc92
|
coiled-coil domain containing 92 |
| chr13_-_79744939 | 0.02 |
ENSRNOT00000076706
|
Suco
|
SUN domain containing ossification factor |
| chr14_-_12387102 | 0.02 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr1_+_217345154 | 0.02 |
ENSRNOT00000092516
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr12_+_41200718 | 0.02 |
ENSRNOT00000038426
ENSRNOT00000048450 ENSRNOT00000067176 |
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr7_+_72924799 | 0.02 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr19_-_25345790 | 0.02 |
ENSRNOT00000010050
|
Zswim4
|
zinc finger, SWIM-type containing 4 |
| chr1_+_84685931 | 0.02 |
ENSRNOT00000063839
|
Zfp780b
|
zinc finger protein 780B |
| chr12_-_50314406 | 0.02 |
ENSRNOT00000079256
|
Hps4
|
Hermansky-Pudlak syndrome 4 |
| chr14_+_84417320 | 0.02 |
ENSRNOT00000008788
|
Tbc1d10a
|
TBC1 domain family, member 10a |
| chr1_-_69743616 | 0.02 |
ENSRNOT00000030259
|
Zfp772
|
zinc finger protein 772 |
| chr5_+_139790395 | 0.02 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr5_-_147828449 | 0.02 |
ENSRNOT00000083369
|
Ccdc28b
|
coiled coil domain containing 28B |
| chr1_-_100234040 | 0.02 |
ENSRNOT00000079199
|
Acpt
|
acid phosphatase, testicular |
| chr1_-_64446818 | 0.02 |
ENSRNOT00000081980
|
Myadm
|
myeloid-associated differentiation marker |
| chr2_+_188141350 | 0.02 |
ENSRNOT00000078913
|
Gon4l
|
gon-4 like |
| chr1_+_140762758 | 0.02 |
ENSRNOT00000047848
|
Acan
|
aggrecan |
| chrX_+_151103576 | 0.02 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr1_+_218569510 | 0.02 |
ENSRNOT00000019652
|
Cpt1a
|
carnitine palmitoyltransferase 1A |
| chr7_+_2827247 | 0.02 |
ENSRNOT00000090689
|
Rnf41
|
ring finger protein 41 |
| chr10_-_70172739 | 0.02 |
ENSRNOT00000010015
|
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr20_+_3156170 | 0.02 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr4_-_157008947 | 0.02 |
ENSRNOT00000073341
|
Pex5
|
peroxisomal biogenesis factor 5 |
| chr2_-_189395746 | 0.02 |
ENSRNOT00000089042
|
Ubap2l
|
ubiquitin associated protein 2-like |
| chr1_+_280383579 | 0.02 |
ENSRNOT00000064022
ENSRNOT00000042977 |
Kcnk18
|
potassium two pore domain channel subfamily K member 18 |
| chr3_+_22854768 | 0.02 |
ENSRNOT00000014630
|
Nek6
|
NIMA-related kinase 6 |
| chr7_+_130474279 | 0.02 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr4_-_156427755 | 0.02 |
ENSRNOT00000085675
ENSRNOT00000014030 |
LOC103690024
|
peroxisomal targeting signal 1 receptor-like |
| chr20_+_48212378 | 0.02 |
ENSRNOT00000040519
|
AABR07045454.1
|
|
| chr1_+_88766872 | 0.02 |
ENSRNOT00000051259
|
Alkbh6
|
alkB homolog 6 |
| chr2_-_210282352 | 0.02 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr8_+_115546893 | 0.02 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr2_-_188216482 | 0.02 |
ENSRNOT00000027579
|
Msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr1_-_212633304 | 0.02 |
ENSRNOT00000025611
|
Olr286
|
olfactory receptor 286 |
| chr9_+_52687868 | 0.02 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr6_+_107428508 | 0.02 |
ENSRNOT00000013431
|
RGD1307704
|
similar to RIKEN cDNA 2410016O06 |
| chr11_-_74315248 | 0.02 |
ENSRNOT00000002346
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr1_+_105285419 | 0.02 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr13_-_35668968 | 0.02 |
ENSRNOT00000042862
ENSRNOT00000003428 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr3_+_164275095 | 0.02 |
ENSRNOT00000077581
|
Snai1
|
snail family transcriptional repressor 1 |
| chr1_-_165967069 | 0.02 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr10_+_15620871 | 0.02 |
ENSRNOT00000089435
ENSRNOT00000066430 |
Nprl3
|
NPR3-like, GATOR1 complex subunit |
| chr4_-_170810080 | 0.02 |
ENSRNOT00000082312
|
Wbp11
|
WW domain binding protein 11 |
| chr1_+_266952561 | 0.02 |
ENSRNOT00000076452
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr7_-_47586137 | 0.02 |
ENSRNOT00000006086
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr9_+_86874685 | 0.02 |
ENSRNOT00000041337
|
NEWGENE_1305560
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr5_-_34813116 | 0.02 |
ENSRNOT00000017479
|
Nkain3
|
Sodium/potassium transporting ATPase interacting 3 |
| chr1_-_72461547 | 0.02 |
ENSRNOT00000022480
|
Ssc5d
|
scavenger receptor cysteine rich family member with 5 domains |
| chr5_-_169160987 | 0.02 |
ENSRNOT00000055487
|
Thap3
|
THAP domain containing 3 |
| chr7_+_118685022 | 0.02 |
ENSRNOT00000089135
|
Apol3
|
apolipoprotein L, 3 |
| chr7_+_38819771 | 0.02 |
ENSRNOT00000006109
|
Lum
|
lumican |
| chr18_-_61057365 | 0.02 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr2_-_197878142 | 0.02 |
ENSRNOT00000087052
|
Tars2
|
threonyl-tRNA synthetase 2, mitochondrial (putative) |
| chr1_-_220188541 | 0.01 |
ENSRNOT00000026968
|
Dpp3
|
dipeptidylpeptidase 3 |
| chrX_+_62727755 | 0.01 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr7_+_3216497 | 0.01 |
ENSRNOT00000008909
|
Mmp19
|
matrix metallopeptidase 19 |
| chr8_-_76579387 | 0.01 |
ENSRNOT00000090747
|
Fam81a
|
family with sequence similarity 81, member A |
| chr7_+_120106901 | 0.01 |
ENSRNOT00000063876
|
Gga1
|
golgi associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr5_-_99033107 | 0.01 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr10_-_88677055 | 0.01 |
ENSRNOT00000025590
|
Ghdc
|
GH3 domain containing |
| chr15_+_52557607 | 0.01 |
ENSRNOT00000019222
|
Gfra2
|
GDNF family receptor alpha 2 |
| chr2_+_189062443 | 0.01 |
ENSRNOT00000028181
|
Adar
|
adenosine deaminase, RNA-specific |
| chr1_-_153740905 | 0.01 |
ENSRNOT00000023239
|
Prss23
|
protease, serine, 23 |
| chr17_+_10384511 | 0.01 |
ENSRNOT00000024357
|
Sncb
|
synuclein, beta |
| chr4_-_117568348 | 0.01 |
ENSRNOT00000071447
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr1_-_165997751 | 0.01 |
ENSRNOT00000050227
|
P2ry6
|
pyrimidinergic receptor P2Y6 |
| chr13_-_110678389 | 0.01 |
ENSRNOT00000082906
|
AABR07022168.1
|
|
| chr10_-_55783489 | 0.01 |
ENSRNOT00000010415
|
Alox15b
|
arachidonate 15-lipoxygenase, type B |
| chr14_-_37871051 | 0.01 |
ENSRNOT00000003087
|
Slain2
|
SLAIN motif family, member 2 |
| chr6_+_136720582 | 0.01 |
ENSRNOT00000086868
|
Kif26a
|
kinesin family member 26A |
| chr8_+_131845696 | 0.01 |
ENSRNOT00000005440
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr16_+_24797124 | 0.01 |
ENSRNOT00000018976
|
Npy5r
|
neuropeptide Y receptor Y5 |
| chr5_-_137125737 | 0.01 |
ENSRNOT00000088532
|
AABR07049768.1
|
|
| chr18_+_63203063 | 0.01 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr14_-_2032593 | 0.01 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr13_-_74331214 | 0.01 |
ENSRNOT00000006018
|
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr7_-_98098268 | 0.01 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr11_-_88177448 | 0.01 |
ENSRNOT00000085475
|
Ypel1
|
yippee-like 1 |
| chrX_+_1311121 | 0.01 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr5_+_157434481 | 0.01 |
ENSRNOT00000088556
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hand1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000041 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.0 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.3 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |