Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
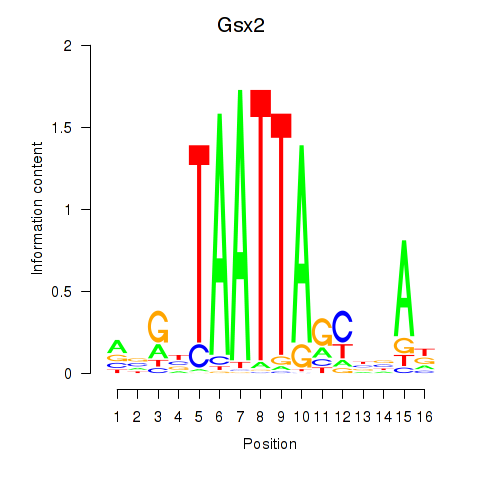
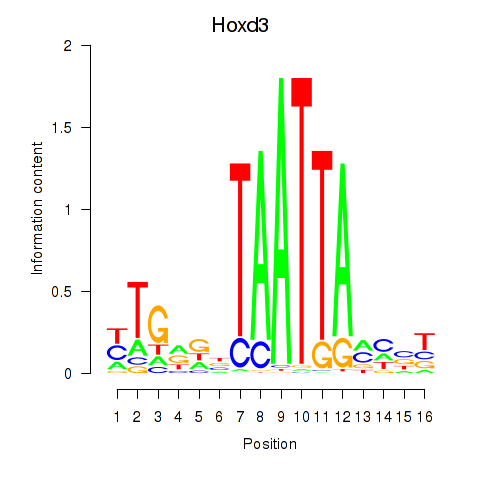
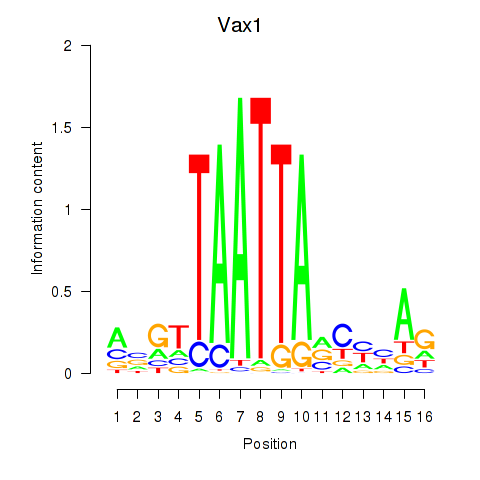
Results for Gsx2_Hoxd3_Vax1
Z-value: 0.36
Transcription factors associated with Gsx2_Hoxd3_Vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx2
|
ENSRNOG00000002266 | GS homeobox 2 |
|
Hoxd3
|
ENSRNOG00000001577 | homeo box D3 |
|
Vax1
|
ENSRNOG00000008824 | ventral anterior homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vax1 | rn6_v1_chr1_-_280338813_280338813 | -0.76 | 1.4e-01 | Click! |
| Hoxd3 | rn6_v1_chr3_+_61658245_61658245 | -0.68 | 2.1e-01 | Click! |
| Gsx2 | rn6_v1_chr14_-_35652709_35652709 | 0.54 | 3.5e-01 | Click! |
Activity profile of Gsx2_Hoxd3_Vax1 motif
Sorted Z-values of Gsx2_Hoxd3_Vax1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_143353925 | 0.31 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr5_+_50381244 | 0.20 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr2_-_219262901 | 0.16 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr8_+_33239139 | 0.13 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr13_+_60619309 | 0.11 |
ENSRNOT00000082129
|
AABR07021204.1
|
|
| chr2_+_66940057 | 0.10 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chrX_-_106607352 | 0.08 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr13_-_89545182 | 0.08 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr4_-_55011415 | 0.08 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr2_-_60657712 | 0.08 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr3_-_90751055 | 0.07 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr2_-_138833933 | 0.07 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr9_-_44419998 | 0.07 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr5_-_17061837 | 0.07 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr8_-_33121002 | 0.07 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr7_-_76488216 | 0.07 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr5_-_17061361 | 0.06 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr3_+_15560712 | 0.06 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_+_71157664 | 0.06 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_-_14364178 | 0.06 |
ENSRNOT00000090673
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chr7_-_101138860 | 0.05 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr6_-_108660063 | 0.05 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr8_+_22559098 | 0.05 |
ENSRNOT00000041091
|
LOC691141
|
hypothetical protein LOC691141 |
| chr20_-_13994794 | 0.05 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr1_+_59156251 | 0.05 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr8_+_22648323 | 0.04 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr8_+_44136496 | 0.04 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr11_-_32550539 | 0.04 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr3_+_72385666 | 0.04 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr8_-_104155775 | 0.04 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr16_+_71889235 | 0.04 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr3_-_145810834 | 0.04 |
ENSRNOT00000075429
|
LOC102555217
|
zinc finger protein 120-like |
| chr2_+_187447501 | 0.04 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr11_-_62067655 | 0.03 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_-_30844199 | 0.03 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr2_+_54466280 | 0.03 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr2_+_252090669 | 0.03 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr7_-_107203897 | 0.03 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr20_-_30947484 | 0.03 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr3_-_165537940 | 0.03 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr11_-_29710849 | 0.03 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr9_+_111279495 | 0.03 |
ENSRNOT00000072791
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr7_-_145062956 | 0.03 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr2_-_149432106 | 0.03 |
ENSRNOT00000051027
|
P2ry13
|
purinergic receptor P2Y13 |
| chr5_-_161981441 | 0.03 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr18_-_69780922 | 0.03 |
ENSRNOT00000079093
|
Me2
|
malic enzyme 2 |
| chr6_-_104290579 | 0.03 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr1_+_48077033 | 0.03 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr3_-_111087347 | 0.03 |
ENSRNOT00000018277
|
Rhov
|
ras homolog family member V |
| chr7_+_133856101 | 0.03 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr7_-_15821927 | 0.03 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr2_+_158097843 | 0.03 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr5_+_187312 | 0.03 |
ENSRNOT00000030857
|
LOC102557117
|
zinc finger protein 120-like |
| chr20_+_3176107 | 0.02 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr2_-_25235275 | 0.02 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr1_+_141767940 | 0.02 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr8_+_59344083 | 0.02 |
ENSRNOT00000031175
|
Crabp1
|
cellular retinoic acid binding protein 1 |
| chr20_-_5166448 | 0.02 |
ENSRNOT00000076331
|
Aif1
|
allograft inflammatory factor 1 |
| chr17_-_43807540 | 0.02 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr19_+_25143428 | 0.02 |
ENSRNOT00000007535
|
Palm3
|
paralemmin 3 |
| chr1_+_80279706 | 0.02 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr17_-_54678710 | 0.02 |
ENSRNOT00000046013
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr1_+_260093641 | 0.02 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr9_-_44237117 | 0.02 |
ENSRNOT00000068496
|
RGD1310819
|
similar to putative protein (5S487) |
| chr7_-_143967484 | 0.02 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr7_-_143497108 | 0.02 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr5_-_133959447 | 0.02 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chrX_+_144994139 | 0.02 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr17_-_84247038 | 0.02 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr2_+_143656793 | 0.02 |
ENSRNOT00000084527
ENSRNOT00000017453 |
Postn
|
periostin |
| chr13_-_82005741 | 0.02 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr6_-_144123596 | 0.02 |
ENSRNOT00000006144
ENSRNOT00000087845 |
Wdr60
|
WD repeat domain 60 |
| chr6_+_29977797 | 0.02 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr1_-_252461461 | 0.02 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr5_-_147412705 | 0.02 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr8_-_84835060 | 0.02 |
ENSRNOT00000007867
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr11_-_14304603 | 0.02 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr3_-_114251647 | 0.02 |
ENSRNOT00000024245
|
Duoxa1
|
dual oxidase maturation factor 1 |
| chr13_-_50916982 | 0.02 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr5_+_64476317 | 0.02 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr3_-_2309179 | 0.02 |
ENSRNOT00000012375
|
Noxa1
|
NADPH oxidase activator 1 |
| chr1_-_276228574 | 0.02 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr1_+_170205591 | 0.02 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr3_+_63510293 | 0.02 |
ENSRNOT00000058093
|
Dfnb59
|
deafness, autosomal recessive 59 |
| chr4_+_31704747 | 0.02 |
ENSRNOT00000047850
|
AABR07059675.1
|
|
| chr8_-_53362006 | 0.02 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr2_+_240642847 | 0.02 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr15_-_11912806 | 0.02 |
ENSRNOT00000068171
ENSRNOT00000008759 ENSRNOT00000049771 |
Slc4a7
|
solute carrier family 4 member 7 |
| chr10_+_94260197 | 0.02 |
ENSRNOT00000063973
|
Taco1
|
translational activator of cytochrome c oxidase I |
| chr1_+_265157379 | 0.01 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr3_-_52664209 | 0.01 |
ENSRNOT00000065126
ENSRNOT00000079020 |
Scn9a
|
sodium voltage-gated channel alpha subunit 9 |
| chr10_+_61685645 | 0.01 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr3_+_112242270 | 0.01 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr12_-_6078411 | 0.01 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr9_+_66058047 | 0.01 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr9_+_61692154 | 0.01 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr17_-_78812111 | 0.01 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr10_-_103619083 | 0.01 |
ENSRNOT00000029092
|
Cd300e
|
Cd300e molecule |
| chr14_-_2032593 | 0.01 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr15_+_45712821 | 0.01 |
ENSRNOT00000083381
ENSRNOT00000045833 |
Fam124a
|
family with sequence similarity 124 member A |
| chr11_+_36851038 | 0.01 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr5_+_142986526 | 0.01 |
ENSRNOT00000012811
|
Rspo1
|
R-spondin 1 |
| chr6_+_122254841 | 0.01 |
ENSRNOT00000005055
|
Gpr65
|
G-protein coupled receptor 65 |
| chr1_-_43638161 | 0.01 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_+_4084228 | 0.01 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chrX_+_78042859 | 0.01 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr2_-_170301348 | 0.01 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr14_-_34218961 | 0.01 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chrX_-_69218526 | 0.01 |
ENSRNOT00000092321
ENSRNOT00000074071 ENSRNOT00000092571 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr4_+_180291389 | 0.01 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr3_-_160561741 | 0.01 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr15_+_33555640 | 0.01 |
ENSRNOT00000021096
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chrX_+_84064427 | 0.01 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr6_+_132510757 | 0.01 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr6_+_18880737 | 0.01 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr7_-_144837583 | 0.01 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr1_+_1825872 | 0.01 |
ENSRNOT00000020417
|
Katna1
|
katanin catalytic subunit A1 |
| chr13_-_82006005 | 0.01 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr9_-_119190698 | 0.01 |
ENSRNOT00000021534
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_+_187988925 | 0.01 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr6_+_104291071 | 0.01 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr1_-_215033460 | 0.01 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr7_+_143122269 | 0.01 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr14_-_6900733 | 0.01 |
ENSRNOT00000061224
|
Dmp1
|
dentin matrix acidic phosphoprotein 1 |
| chr9_+_67234303 | 0.01 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr8_-_40137390 | 0.01 |
ENSRNOT00000042717
|
Panx3
|
pannexin 3 |
| chr18_-_16543992 | 0.01 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr20_+_3246739 | 0.01 |
ENSRNOT00000061299
|
RT1-T24-2
|
RT1 class I, locus T24, gene 2 |
| chr3_+_72395218 | 0.01 |
ENSRNOT00000057616
|
Prg3
|
proteoglycan 3, pro eosinophil major basic protein 2 |
| chr10_-_88000423 | 0.01 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr15_-_28517335 | 0.01 |
ENSRNOT00000060514
ENSRNOT00000090378 |
LOC100911576
|
heterogeneous nuclear ribonucleoproteins C1/C2-like |
| chrX_+_68627313 | 0.01 |
ENSRNOT00000076699
ENSRNOT00000076795 ENSRNOT00000008705 |
Yipf6
|
Yip1 domain family, member 6 |
| chr18_+_30036887 | 0.01 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr8_+_70447040 | 0.01 |
ENSRNOT00000016798
|
Ints14
|
integrator complex subunit 14 |
| chr8_+_2604962 | 0.01 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr4_-_72143748 | 0.01 |
ENSRNOT00000024428
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr1_-_256734719 | 0.01 |
ENSRNOT00000021546
ENSRNOT00000089456 |
Myof
|
myoferlin |
| chr3_-_165700489 | 0.01 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chrX_-_152642531 | 0.01 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr9_-_85243001 | 0.01 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr12_+_30441055 | 0.01 |
ENSRNOT00000081099
|
AABR07036007.1
|
|
| chr6_+_104291340 | 0.01 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr1_-_63293635 | 0.01 |
ENSRNOT00000090337
|
Zik1
|
zinc finger protein interacting with K protein 1 |
| chr11_-_61234944 | 0.01 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr9_+_10013854 | 0.01 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr3_-_15376767 | 0.01 |
ENSRNOT00000044885
|
Ndufa8
|
NADH:ubiquinone oxidoreductase subunit A8 |
| chr16_+_20825044 | 0.01 |
ENSRNOT00000027268
|
Upf1
|
UPF1, RNA helicase and ATPase |
| chrX_+_112311251 | 0.01 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr18_+_17043903 | 0.01 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr3_-_14643897 | 0.01 |
ENSRNOT00000082008
ENSRNOT00000025983 |
Ggta1
|
glycoprotein, alpha-galactosyltransferase 1 |
| chr1_-_23556241 | 0.01 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr6_+_137323713 | 0.01 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr20_+_3230052 | 0.01 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr2_+_202200797 | 0.01 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr2_-_93985378 | 0.01 |
ENSRNOT00000075493
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr9_+_65614142 | 0.01 |
ENSRNOT00000016613
|
Casp8
|
caspase 8 |
| chr4_+_6827429 | 0.01 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr9_+_60883981 | 0.01 |
ENSRNOT00000081002
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr5_+_18901039 | 0.01 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr17_+_55065621 | 0.01 |
ENSRNOT00000086342
|
Zfp438
|
zinc finger protein 438 |
| chr4_+_153805993 | 0.00 |
ENSRNOT00000056174
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr2_+_19823234 | 0.00 |
ENSRNOT00000022348
|
Rps23
|
ribosomal protein S23 |
| chrX_-_70978952 | 0.00 |
ENSRNOT00000076910
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr15_+_108526014 | 0.00 |
ENSRNOT00000017211
|
Tm9sf2
|
transmembrane 9 superfamily member 2 |
| chr2_-_250862419 | 0.00 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr7_-_55604403 | 0.00 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr4_+_158088505 | 0.00 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr4_+_24612205 | 0.00 |
ENSRNOT00000057382
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr14_+_39964588 | 0.00 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr13_+_76962504 | 0.00 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr20_-_13458110 | 0.00 |
ENSRNOT00000072183
|
Slc5a4
|
solute carrier family 5 member 4 |
| chr16_+_74865516 | 0.00 |
ENSRNOT00000058072
|
Atp7b
|
ATPase copper transporting beta |
| chr13_-_78885464 | 0.00 |
ENSRNOT00000003828
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr2_-_187113717 | 0.00 |
ENSRNOT00000020147
|
Lrrc71
|
leucine rich repeat containing 71 |
| chr1_+_229416489 | 0.00 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chr10_-_71491743 | 0.00 |
ENSRNOT00000038955
|
LOC102552988
|
uncharacterized LOC102552988 |
| chr1_-_221420115 | 0.00 |
ENSRNOT00000028475
|
Mrpl49
|
mitochondrial ribosomal protein L49 |
| chr18_+_32336102 | 0.00 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr15_-_54528480 | 0.00 |
ENSRNOT00000066888
|
Fndc3a
|
fibronectin type III domain containing 3a |
| chr3_+_72134731 | 0.00 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr8_+_103938520 | 0.00 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr1_-_81550598 | 0.00 |
ENSRNOT00000051671
|
RGD1564380
|
similar to BC049730 protein |
| chr6_+_95205153 | 0.00 |
ENSRNOT00000007339
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr10_+_88227360 | 0.00 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr8_-_17525906 | 0.00 |
ENSRNOT00000007855
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr17_+_89951752 | 0.00 |
ENSRNOT00000057572
ENSRNOT00000091023 |
Abi1
|
abl-interactor 1 |
| chrX_+_66314418 | 0.00 |
ENSRNOT00000035405
|
Pgr15l
|
G protein-coupled receptor 15-like |
| chr2_+_266315036 | 0.00 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr8_-_78233430 | 0.00 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr20_+_13817795 | 0.00 |
ENSRNOT00000036518
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr7_+_2504695 | 0.00 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr3_-_3594475 | 0.00 |
ENSRNOT00000064861
|
RGD1564379
|
RGD1564379 |
| chr12_+_2140203 | 0.00 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chrM_+_5323 | 0.00 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr16_+_60925093 | 0.00 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chrM_+_9870 | 0.00 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chrX_+_14019961 | 0.00 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr6_-_127319362 | 0.00 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr12_+_17253791 | 0.00 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr13_+_47935163 | 0.00 |
ENSRNOT00000006768
|
Eif2d
|
eukaryotic translation initiation factor 2D |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx2_Hoxd3_Vax1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |