Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
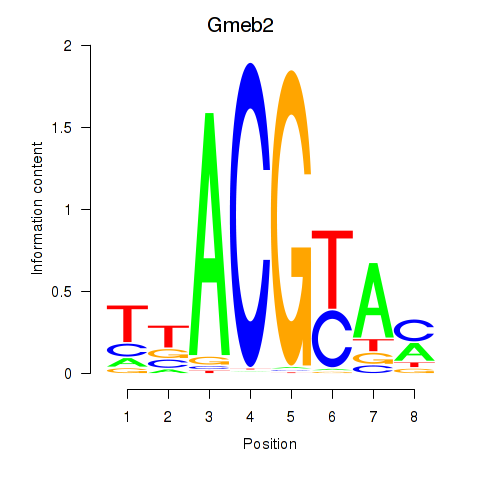
Results for Gmeb2
Z-value: 0.12
Transcription factors associated with Gmeb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gmeb2
|
ENSRNOG00000013339 | glucocorticoid modulatory element binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gmeb2 | rn6_v1_chr3_-_176781040_176781040 | 0.70 | 1.9e-01 | Click! |
Activity profile of Gmeb2 motif
Sorted Z-values of Gmeb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26390689 | 0.04 |
ENSRNOT00000079762
ENSRNOT00000091528 |
Ift172
|
intraflagellar transport 172 |
| chr7_-_141307233 | 0.03 |
ENSRNOT00000071885
|
Racgap1
|
Rac GTPase-activating protein 1 |
| chr14_+_42714315 | 0.03 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr6_+_8346704 | 0.02 |
ENSRNOT00000092218
|
AC111831.2
|
|
| chr1_-_190370499 | 0.02 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr11_+_47243342 | 0.02 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr5_+_57291156 | 0.02 |
ENSRNOT00000060692
ENSRNOT00000060691 |
Nfx1
|
nuclear transcription factor, X-box binding 1 |
| chr7_-_119768082 | 0.02 |
ENSRNOT00000009612
|
Sstr3
|
somatostatin receptor 3 |
| chr10_+_90230441 | 0.02 |
ENSRNOT00000082722
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr19_+_55300395 | 0.02 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr19_+_16415636 | 0.02 |
ENSRNOT00000089975
|
Irx5
|
iroquois homeobox 5 |
| chr3_+_33641616 | 0.02 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr5_+_148320438 | 0.01 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr4_+_199916 | 0.01 |
ENSRNOT00000009317
|
Htr5a
|
5-hydroxytryptamine receptor 5A |
| chr4_-_176789304 | 0.01 |
ENSRNOT00000018057
|
Kcnj8
|
potassium voltage-gated channel subfamily J member 8 |
| chr19_+_37668693 | 0.01 |
ENSRNOT00000084970
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr13_-_73704480 | 0.01 |
ENSRNOT00000005296
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr14_+_79261092 | 0.01 |
ENSRNOT00000029191
|
LOC680039
|
hypothetical protein LOC680039 |
| chr3_+_164986421 | 0.01 |
ENSRNOT00000039403
|
Mocs3
|
molybdenum cofactor synthesis 3 |
| chr10_-_71837851 | 0.01 |
ENSRNOT00000000258
|
Aatf
|
apoptosis antagonizing transcription factor |
| chr5_+_22392732 | 0.01 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr9_+_88110731 | 0.01 |
ENSRNOT00000088677
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr11_-_71419223 | 0.01 |
ENSRNOT00000002407
|
Tfrc
|
transferrin receptor |
| chr8_+_79660657 | 0.01 |
ENSRNOT00000090970
|
Ccpg1
|
cell cycle progression 1 |
| chr10_-_56289882 | 0.01 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr9_+_10428853 | 0.01 |
ENSRNOT00000074253
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chrX_-_105417323 | 0.01 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr14_+_84211600 | 0.01 |
ENSRNOT00000005996
|
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr5_+_122100099 | 0.01 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr7_+_29282305 | 0.01 |
ENSRNOT00000007623
|
Arl1
|
ADP-ribosylation factor like GTPase 1 |
| chr18_-_57515834 | 0.01 |
ENSRNOT00000026098
|
Adrb2
|
adrenoceptor beta 2 |
| chr20_-_27308069 | 0.01 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr6_-_77243677 | 0.01 |
ENSRNOT00000011419
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr12_+_37444072 | 0.01 |
ENSRNOT00000077380
ENSRNOT00000092699 ENSRNOT00000001373 ENSRNOT00000092416 |
Eif2b1
|
eukaryotic translation initiation factor 2B subunit 1 alpha |
| chr1_-_199865548 | 0.01 |
ENSRNOT00000027463
|
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr1_+_154377447 | 0.01 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr18_-_27749235 | 0.01 |
ENSRNOT00000026696
|
Hspa9
|
heat shock protein family A member 9 |
| chr20_+_28027054 | 0.01 |
ENSRNOT00000071386
ENSRNOT00000001044 |
Ranbp2
|
RAN binding protein 2 |
| chr6_+_52751106 | 0.01 |
ENSRNOT00000014373
|
Twistnb
|
TWIST neighbor |
| chr1_+_167538263 | 0.01 |
ENSRNOT00000074058
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr12_-_50383496 | 0.01 |
ENSRNOT00000000831
|
Tpst2
|
tyrosylprotein sulfotransferase 2 |
| chr6_-_50943488 | 0.01 |
ENSRNOT00000068419
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr1_-_189666440 | 0.01 |
ENSRNOT00000019167
|
Dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr8_+_104040934 | 0.01 |
ENSRNOT00000081204
|
Tfdp2
|
transcription factor Dp-2 |
| chr5_+_144779681 | 0.00 |
ENSRNOT00000016493
|
LOC100294508
|
dyslexia susceptibility 2-like |
| chr5_-_151072766 | 0.00 |
ENSRNOT00000078048
ENSRNOT00000017227 ENSRNOT00000085573 |
Stx12
|
syntaxin 12 |
| chr2_-_196456481 | 0.00 |
ENSRNOT00000028677
|
Prune
|
prune exopolyphosphatase |
| chr8_+_117280705 | 0.00 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr20_-_32115310 | 0.00 |
ENSRNOT00000067883
ENSRNOT00000081916 |
Vps26a
|
VPS26 retromer complex component A |
| chr12_+_11179329 | 0.00 |
ENSRNOT00000001302
|
Zfp394
|
zinc finger protein 394 |
| chr14_-_86706626 | 0.00 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr2_-_232178533 | 0.00 |
ENSRNOT00000082173
ENSRNOT00000055607 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr17_+_26808330 | 0.00 |
ENSRNOT00000022108
|
Bloc1s5
|
biogenesis of lysosomal organelles complex 1 subunit 5 |
| chr7_-_29282039 | 0.00 |
ENSRNOT00000067777
|
RGD1561102
|
similar to ribosomal protein S12 |
| chr5_+_157848206 | 0.00 |
ENSRNOT00000039663
ENSRNOT00000082138 |
Ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr5_-_173098816 | 0.00 |
ENSRNOT00000023696
|
Mib2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr20_-_5455632 | 0.00 |
ENSRNOT00000000552
|
Wdr46
|
WD repeat domain 46 |
| chr15_-_19733967 | 0.00 |
ENSRNOT00000012036
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr1_+_127802978 | 0.00 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr7_-_120780108 | 0.00 |
ENSRNOT00000018185
|
Ddx17
|
DEAD-box helicase 17 |
| chr6_+_96871625 | 0.00 |
ENSRNOT00000012361
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr10_-_38642397 | 0.00 |
ENSRNOT00000023628
|
Hspa4
|
heat shock protein family A member 4 |
| chr10_+_86724427 | 0.00 |
ENSRNOT00000013204
|
Casc3
|
cancer susceptibility candidate 3 |
| chr10_+_90230711 | 0.00 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr10_+_83636518 | 0.00 |
ENSRNOT00000007352
|
Phospho1
|
phosphoethanolamine/phosphocholine phosphatase 1 |
| chr10_+_97647111 | 0.00 |
ENSRNOT00000055062
|
Gna13
|
G protein subunit alpha 13 |
| chr14_+_2100106 | 0.00 |
ENSRNOT00000000064
|
Gak
|
cyclin G associated kinase |
| chr8_+_61080103 | 0.00 |
ENSRNOT00000022976
|
Hmg20a
|
high mobility group 20A |
| chr1_+_189665976 | 0.00 |
ENSRNOT00000033673
|
Lyrm1
|
LYR motif containing 1 |
| chr9_+_114022137 | 0.00 |
ENSRNOT00000007657
|
Map3k7
|
mitogen activated protein kinase kinase kinase 7 |
| chr20_-_32080088 | 0.00 |
ENSRNOT00000080773
|
Supv3l1
|
Suv3 like RNA helicase |
| chr3_-_133131192 | 0.00 |
ENSRNOT00000055630
|
Tasp1
|
taspase 1 |
| chrX_+_159703578 | 0.00 |
ENSRNOT00000001162
|
Cd40lg
|
CD40 ligand |
| chr7_-_120780641 | 0.00 |
ENSRNOT00000076164
|
Ddx17
|
DEAD-box helicase 17 |
| chr8_+_116054465 | 0.00 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr11_-_31805728 | 0.00 |
ENSRNOT00000032162
|
Gart
|
phosphoribosylglycinamide formyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gmeb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |