Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
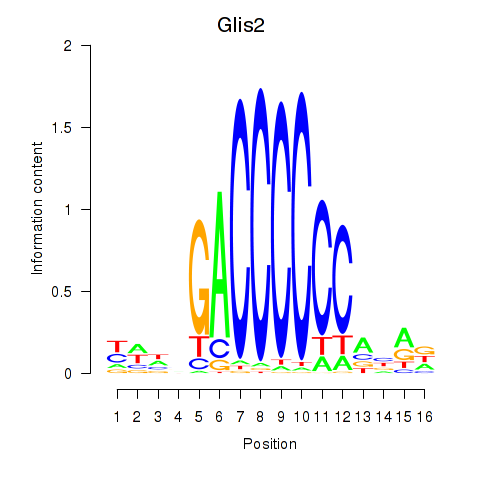
Results for Glis2
Z-value: 0.32
Transcription factors associated with Glis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis2
|
ENSRNOG00000004766 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis2 | rn6_v1_chr10_-_11174861_11174861 | 0.07 | 9.1e-01 | Click! |
Activity profile of Glis2 motif
Sorted Z-values of Glis2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_123445613 | 0.37 |
ENSRNOT00000070937
|
Shisa8
|
shisa family member 8 |
| chr10_-_88551056 | 0.15 |
ENSRNOT00000032737
|
Zfp385c
|
zinc finger protein 385C |
| chr12_-_38782010 | 0.14 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr5_-_151709877 | 0.12 |
ENSRNOT00000080602
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr7_+_11737293 | 0.09 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr17_-_1093873 | 0.09 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr7_-_28040510 | 0.08 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr16_-_50501716 | 0.07 |
ENSRNOT00000067486
|
Fat1
|
FAT atypical cadherin 1 |
| chr10_+_72486152 | 0.07 |
ENSRNOT00000004193
|
RGD1310166
|
similar to Chromodomain-helicase-DNA-binding protein 1 (CHD-1) |
| chr14_-_35149608 | 0.07 |
ENSRNOT00000003050
ENSRNOT00000090654 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr10_+_91710495 | 0.06 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr1_-_87468288 | 0.06 |
ENSRNOT00000042207
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr10_+_40553180 | 0.06 |
ENSRNOT00000087763
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr10_-_85938085 | 0.06 |
ENSRNOT00000043561
|
Plxdc1
|
plexin domain containing 1 |
| chr4_-_123494742 | 0.06 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr3_+_148541909 | 0.05 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr10_+_61403130 | 0.05 |
ENSRNOT00000092490
ENSRNOT00000077649 |
Ccdc92b
|
coiled-coil domain containing 92B |
| chr12_-_47793534 | 0.05 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr10_-_82197520 | 0.05 |
ENSRNOT00000092024
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr7_-_120490061 | 0.05 |
ENSRNOT00000016247
|
Slc16a8
|
solute carrier family 16 member 8 |
| chr14_+_34389991 | 0.05 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr15_+_57505449 | 0.05 |
ENSRNOT00000064945
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr3_+_148579920 | 0.05 |
ENSRNOT00000012432
|
Hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr12_+_2046472 | 0.05 |
ENSRNOT00000001289
|
Zfp358
|
zinc finger protein 358 |
| chr15_+_34520142 | 0.05 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr17_+_80882666 | 0.05 |
ENSRNOT00000024430
|
Vim
|
vimentin |
| chr12_+_9360672 | 0.05 |
ENSRNOT00000088957
|
Flt3
|
fms-related tyrosine kinase 3 |
| chr1_-_219719272 | 0.05 |
ENSRNOT00000026152
|
Syt12
|
synaptotagmin 12 |
| chr1_-_101596822 | 0.05 |
ENSRNOT00000028490
|
Fgf21
|
fibroblast growth factor 21 |
| chr1_-_184188911 | 0.04 |
ENSRNOT00000055124
ENSRNOT00000014948 |
Calca
|
calcitonin-related polypeptide alpha |
| chr6_-_107325345 | 0.04 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr10_+_56381813 | 0.04 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr11_+_60073383 | 0.04 |
ENSRNOT00000087508
|
Tagln3
|
transgelin 3 |
| chr8_+_130581062 | 0.04 |
ENSRNOT00000014323
|
Fam198a
|
family with sequence similarity 198, member A |
| chr11_+_57505005 | 0.04 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr3_-_72289310 | 0.04 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr16_+_2743823 | 0.04 |
ENSRNOT00000087873
|
Arhgef3
|
Rho guanine nucleotide exchange factor 3 |
| chr1_+_124625985 | 0.04 |
ENSRNOT00000021068
|
Otud7a
|
OTU deubiquitinase 7A |
| chr9_+_16702460 | 0.04 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chr1_+_177193735 | 0.03 |
ENSRNOT00000043017
|
Micalcl
|
MICAL C-terminal like |
| chr1_-_164308317 | 0.03 |
ENSRNOT00000022983
|
Serpinh1
|
serpin family H member 1 |
| chr15_+_40665041 | 0.03 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr9_-_79545024 | 0.03 |
ENSRNOT00000021152
|
Mreg
|
melanoregulin |
| chr18_-_71614980 | 0.03 |
ENSRNOT00000032563
|
LOC102548286
|
peroxisomal biogenesis factor 19-like |
| chr7_+_70556019 | 0.03 |
ENSRNOT00000087495
ENSRNOT00000008120 |
Dctn2
|
dynactin subunit 2 |
| chr14_-_28856214 | 0.03 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr12_+_25119355 | 0.03 |
ENSRNOT00000034629
|
Lat2
|
linker for activation of T cells family, member 2 |
| chr3_-_143063983 | 0.03 |
ENSRNOT00000006329
|
Napb
|
NSF attachment protein beta |
| chr7_-_12311149 | 0.03 |
ENSRNOT00000046886
|
Dazap1
|
DAZ associated protein 1 |
| chr17_+_23135985 | 0.03 |
ENSRNOT00000090794
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr5_-_58078545 | 0.03 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr7_-_129724303 | 0.03 |
ENSRNOT00000006034
|
Brd1
|
bromodomain containing 1 |
| chr12_+_15857805 | 0.03 |
ENSRNOT00000001667
|
Iqce
|
IQ motif containing E |
| chr9_-_81565416 | 0.03 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr10_-_70202398 | 0.03 |
ENSRNOT00000076089
|
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr17_-_44840131 | 0.02 |
ENSRNOT00000083417
|
Hist1h3b
|
histone cluster 1, H3b |
| chr3_-_148407778 | 0.02 |
ENSRNOT00000011262
|
Foxs1
|
forkhead box S1 |
| chr1_+_100832324 | 0.02 |
ENSRNOT00000056364
|
Il4i1
|
interleukin 4 induced 1 |
| chr8_-_13109487 | 0.02 |
ENSRNOT00000061616
|
Amotl1
|
angiomotin-like 1 |
| chr8_+_116754178 | 0.02 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr11_-_89260297 | 0.02 |
ENSRNOT00000057502
|
Spidr
|
scaffolding protein involved in DNA repair |
| chr1_-_127292090 | 0.02 |
ENSRNOT00000016959
|
Lrrk1
|
leucine-rich repeat kinase 1 |
| chr20_-_4508197 | 0.02 |
ENSRNOT00000086027
ENSRNOT00000000514 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr9_-_110936631 | 0.02 |
ENSRNOT00000064431
|
Fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chr1_+_170471238 | 0.02 |
ENSRNOT00000076961
ENSRNOT00000075597 ENSRNOT00000076631 ENSRNOT00000076783 |
Timm10b
Dnhd1
|
translocase of inner mitochondrial membrane 10B dynein heavy chain domain 1 |
| chr5_-_150925727 | 0.02 |
ENSRNOT00000017618
|
Xkr8
|
XK related 8 |
| chr15_-_38276159 | 0.02 |
ENSRNOT00000084698
ENSRNOT00000015233 |
Micu2
|
mitochondrial calcium uptake 2 |
| chr6_+_123897956 | 0.02 |
ENSRNOT00000005167
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr14_+_85250042 | 0.02 |
ENSRNOT00000057407
|
Ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr20_-_1980101 | 0.02 |
ENSRNOT00000084582
ENSRNOT00000085050 ENSRNOT00000082545 ENSRNOT00000088396 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr10_+_107455845 | 0.02 |
ENSRNOT00000004373
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_-_119768082 | 0.02 |
ENSRNOT00000009612
|
Sstr3
|
somatostatin receptor 3 |
| chr1_+_199196059 | 0.02 |
ENSRNOT00000090428
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr3_+_80556668 | 0.02 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr16_-_18860086 | 0.02 |
ENSRNOT00000074814
|
Sin3b
|
SIN3 transcription regulator family member B |
| chr2_-_164126783 | 0.02 |
ENSRNOT00000016843
|
Shox2
|
short stature homeobox 2 |
| chr11_+_60072727 | 0.02 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr3_-_148312791 | 0.02 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr13_+_90514336 | 0.02 |
ENSRNOT00000088996
ENSRNOT00000085377 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr12_-_13157570 | 0.02 |
ENSRNOT00000001437
ENSRNOT00000067332 ENSRNOT00000092238 |
Daglb
|
diacylglycerol lipase, beta |
| chr20_+_28572242 | 0.02 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr19_+_37226186 | 0.02 |
ENSRNOT00000075933
ENSRNOT00000065013 |
Hsf4
|
heat shock transcription factor 4 |
| chr1_-_220502272 | 0.02 |
ENSRNOT00000041238
|
Klc2
|
kinesin light chain 2 |
| chr2_+_62360754 | 0.01 |
ENSRNOT00000017276
|
Golph3
|
golgi phosphoprotein 3 |
| chrX_+_65566047 | 0.01 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr10_-_57436368 | 0.01 |
ENSRNOT00000056608
|
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr18_-_24057917 | 0.01 |
ENSRNOT00000023874
|
Rit2
|
Ras-like without CAAX 2 |
| chr20_-_2212171 | 0.01 |
ENSRNOT00000085912
|
Trim26
|
tripartite motif-containing 26 |
| chr8_+_133197032 | 0.01 |
ENSRNOT00000075564
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr20_+_11114164 | 0.01 |
ENSRNOT00000001602
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr3_+_149424392 | 0.01 |
ENSRNOT00000044574
|
Bpifb4
|
BPI fold containing family B, member 4 |
| chr20_-_2211995 | 0.01 |
ENSRNOT00000077201
|
Trim26
|
tripartite motif-containing 26 |
| chrX_-_123515720 | 0.01 |
ENSRNOT00000092343
|
Nkrf
|
NFKB repressing factor |
| chr11_-_88273254 | 0.01 |
ENSRNOT00000002533
|
Mapk1
|
mitogen activated protein kinase 1 |
| chr2_-_198719202 | 0.01 |
ENSRNOT00000028801
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr13_-_89594738 | 0.01 |
ENSRNOT00000004641
|
Tomm40l
|
translocase of outer mitochondrial membrane 40 like |
| chr1_+_82480195 | 0.01 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr10_-_37311625 | 0.01 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr7_+_70328391 | 0.01 |
ENSRNOT00000075544
|
Mettl1
|
methyltransferase like 1 |
| chr1_+_123015746 | 0.01 |
ENSRNOT00000013483
|
Magel2
|
MAGE family member L2 |
| chr5_-_138470096 | 0.01 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr3_-_111469518 | 0.01 |
ENSRNOT00000006874
|
Ndufaf1
|
NADH:ubiquinone oxidoreductase complex assembly factor 1 |
| chr12_+_22466601 | 0.01 |
ENSRNOT00000074544
|
AABR07035787.1
|
|
| chr20_-_10257044 | 0.01 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr3_+_111160205 | 0.01 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr18_+_15738809 | 0.01 |
ENSRNOT00000075444
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_+_70364813 | 0.01 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr10_+_43538160 | 0.01 |
ENSRNOT00000089217
|
Cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chrX_+_65226748 | 0.01 |
ENSRNOT00000076181
|
Msn
|
moesin |
| chr12_+_25036605 | 0.01 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chrX_+_71324365 | 0.01 |
ENSRNOT00000004911
|
Nono
|
non-POU domain containing, octamer-binding |
| chr2_+_196139755 | 0.01 |
ENSRNOT00000040501
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chrX_+_5825711 | 0.01 |
ENSRNOT00000037360
|
Efhc2
|
EF-hand domain containing 2 |
| chr16_-_7007287 | 0.01 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr4_+_147333056 | 0.01 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr11_+_45751812 | 0.01 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr16_-_23156962 | 0.01 |
ENSRNOT00000018543
|
Sh2d4a
|
SH2 domain containing 4A |
| chr1_-_219853329 | 0.01 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr10_+_67494984 | 0.01 |
ENSRNOT00000056273
|
Adap2
|
ArfGAP with dual PH domains 2 |
| chr10_+_90984227 | 0.01 |
ENSRNOT00000003890
ENSRNOT00000093707 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr1_+_85162452 | 0.01 |
ENSRNOT00000093347
|
Pak4
|
p21 (RAC1) activated kinase 4 |
| chr9_+_81656116 | 0.01 |
ENSRNOT00000083421
|
Slc11a1
|
solute carrier family 11 member 1 |
| chr2_-_178057157 | 0.00 |
ENSRNOT00000091135
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr20_+_30791422 | 0.00 |
ENSRNOT00000047394
ENSRNOT00000000683 |
Tbata
|
thymus, brain and testes associated |
| chr8_+_21890319 | 0.00 |
ENSRNOT00000027949
|
LOC500956
|
hypothetical protein LOC500956 |
| chr20_-_2678141 | 0.00 |
ENSRNOT00000072377
ENSRNOT00000083833 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr12_+_23544287 | 0.00 |
ENSRNOT00000001938
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr12_-_29958050 | 0.00 |
ENSRNOT00000058725
|
Tmem248
|
transmembrane protein 248 |
| chr15_+_34234193 | 0.00 |
ENSRNOT00000077584
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr5_+_135574172 | 0.00 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr7_+_143629455 | 0.00 |
ENSRNOT00000073951
|
Krt18
|
keratin 18 |
| chr2_-_188645196 | 0.00 |
ENSRNOT00000083793
|
Efna3
|
ephrin A3 |
| chr6_+_136084605 | 0.00 |
ENSRNOT00000092806
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr3_+_2506426 | 0.00 |
ENSRNOT00000015242
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr4_+_27175243 | 0.00 |
ENSRNOT00000009985
|
Cyp51
|
cytochrome P450, family 51 |
| chr4_-_7885301 | 0.00 |
ENSRNOT00000035034
|
Rint1
|
RAD50 interactor 1 |
| chr20_-_5441706 | 0.00 |
ENSRNOT00000000549
|
Vps52
|
VPS52 GARP complex subunit |
| chr3_+_172385672 | 0.00 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr4_+_161685258 | 0.00 |
ENSRNOT00000008012
ENSRNOT00000008003 |
Foxm1
|
forkhead box M1 |
| chr3_+_11756384 | 0.00 |
ENSRNOT00000087762
|
Sh2d3c
|
SH2 domain containing 3C |
| chr7_-_124317379 | 0.00 |
ENSRNOT00000013398
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr8_+_117574161 | 0.00 |
ENSRNOT00000077078
|
Ip6k2
|
inositol hexakisphosphate kinase 2 |
| chr1_+_16819170 | 0.00 |
ENSRNOT00000019734
ENSRNOT00000086756 |
Hbs1l
|
HBS1-like translational GTPase |
| chr10_+_88490798 | 0.00 |
ENSRNOT00000023872
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr4_-_157155609 | 0.00 |
ENSRNOT00000016330
|
C1s
|
complement C1s |
| chr19_+_37305248 | 0.00 |
ENSRNOT00000068299
|
Slc9a5
|
solute carrier family 9 member A5 |
| chr1_+_72425707 | 0.00 |
ENSRNOT00000058956
|
Sbk2
|
SH3 domain binding kinase family, member 2 |
| chr6_-_25507073 | 0.00 |
ENSRNOT00000061176
|
Plb1
|
phospholipase B1 |
| chr1_-_80580662 | 0.00 |
ENSRNOT00000024779
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr16_+_6970342 | 0.00 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr2_+_250600823 | 0.00 |
ENSRNOT00000083750
|
Sep15
|
selenoprotein 15 |
| chr14_+_21972274 | 0.00 |
ENSRNOT00000060265
|
Csn2
|
casein beta |
| chr7_-_144269486 | 0.00 |
ENSRNOT00000090051
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr8_-_116532169 | 0.00 |
ENSRNOT00000085364
|
Rbm5
|
RNA binding motif protein 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0021550 | neuroblast differentiation(GO:0014016) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0097324 | Kit signaling pathway(GO:0038109) melanocyte migration(GO:0097324) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |