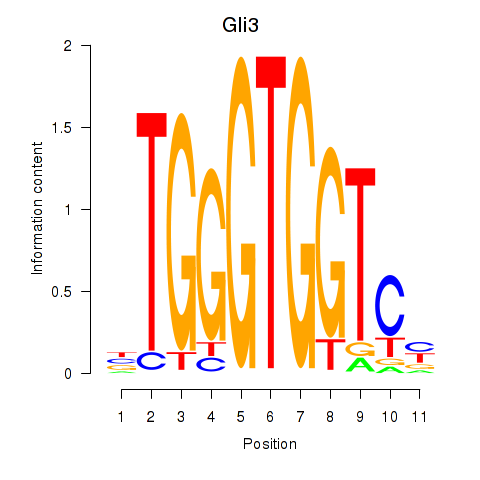
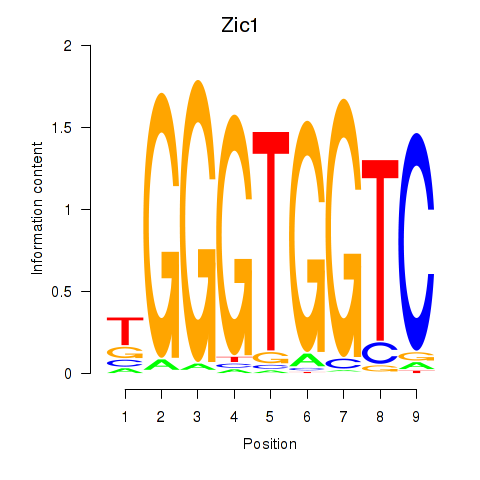
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Gli3_Zic1
Z-value: 0.60
Transcription factors associated with Gli3_Zic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli3
|
ENSRNOG00000014395 | GLI family zinc finger 3 |
|
Zic1
|
ENSRNOG00000014644 | Zic family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli3 | rn6_v1_chr17_-_52477575_52477575 | -0.84 | 7.4e-02 | Click! |
| Zic1 | rn6_v1_chr8_-_98738446_98738446 | 0.36 | 5.5e-01 | Click! |
Activity profile of Gli3_Zic1 motif
Sorted Z-values of Gli3_Zic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_90312386 | 0.44 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr1_-_216080287 | 0.39 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr6_-_27190126 | 0.34 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr1_-_80744831 | 0.31 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr3_+_154043873 | 0.20 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr18_-_28454756 | 0.19 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr5_-_77945016 | 0.18 |
ENSRNOT00000076223
|
Zfp37
|
zinc finger protein 37 |
| chr16_+_54332660 | 0.17 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr10_+_91710495 | 0.16 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr7_-_12326392 | 0.14 |
ENSRNOT00000039728
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chr4_+_24617008 | 0.12 |
ENSRNOT00000086607
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr12_+_47074200 | 0.12 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr6_+_34155017 | 0.12 |
ENSRNOT00000073663
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr1_-_259357056 | 0.12 |
ENSRNOT00000022000
|
Pdlim1
|
PDZ and LIM domain 1 |
| chr4_+_153774486 | 0.11 |
ENSRNOT00000074096
|
Tuba8
|
tubulin, alpha 8 |
| chr1_-_72366455 | 0.11 |
ENSRNOT00000031786
|
LOC100911196
|
zinc finger protein 865-like |
| chr9_+_80118029 | 0.11 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr12_+_2046472 | 0.11 |
ENSRNOT00000001289
|
Zfp358
|
zinc finger protein 358 |
| chr4_+_55715744 | 0.11 |
ENSRNOT00000010429
|
Arf5
|
ADP-ribosylation factor 5 |
| chr8_-_55177818 | 0.10 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr6_+_137997335 | 0.10 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr16_-_82439441 | 0.10 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr5_-_17061361 | 0.10 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr4_+_117962319 | 0.10 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chrX_-_13116743 | 0.09 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr10_-_108196217 | 0.09 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr7_+_11769400 | 0.09 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr8_+_65686648 | 0.09 |
ENSRNOT00000017233
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr10_+_46314375 | 0.09 |
ENSRNOT00000088300
|
Med9
|
mediator complex subunit 9 |
| chr16_+_21288876 | 0.09 |
ENSRNOT00000027990
|
Cilp2
|
cartilage intermediate layer protein 2 |
| chr18_+_32594958 | 0.09 |
ENSRNOT00000018511
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr1_+_101783621 | 0.08 |
ENSRNOT00000067679
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr16_+_80826681 | 0.08 |
ENSRNOT00000000123
|
Coprs
|
coordinator of PRMT5 and differentiation stimulator |
| chrX_-_106747303 | 0.08 |
ENSRNOT00000073529
|
Tceal5
|
transcription elongation factor A like 5 |
| chr5_+_135962911 | 0.08 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chr1_+_207508414 | 0.07 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr19_-_37938857 | 0.07 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr2_-_235831670 | 0.07 |
ENSRNOT00000037393
|
Ostc
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr9_+_82556573 | 0.07 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr4_-_100783016 | 0.07 |
ENSRNOT00000020671
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr17_-_1093873 | 0.07 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr2_-_172459165 | 0.06 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_-_176607466 | 0.06 |
ENSRNOT00000022984
|
Galnt18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr6_+_132702448 | 0.06 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr11_-_25350974 | 0.06 |
ENSRNOT00000002187
|
Adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr12_+_39790965 | 0.06 |
ENSRNOT00000030604
|
Tctn1
|
tectonic family member 1 |
| chr1_+_97632473 | 0.06 |
ENSRNOT00000023671
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr10_+_65448950 | 0.06 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr5_-_60871705 | 0.06 |
ENSRNOT00000016582
|
Exosc3
|
exosome component 3 |
| chr9_-_82699551 | 0.06 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr1_+_81208212 | 0.06 |
ENSRNOT00000026288
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr9_+_43331155 | 0.06 |
ENSRNOT00000023036
|
Zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr15_+_2374582 | 0.06 |
ENSRNOT00000019644
|
Zfp503
|
zinc finger protein 503 |
| chr6_+_51662224 | 0.06 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr10_+_86860685 | 0.06 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr1_+_84304228 | 0.06 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr2_-_115836846 | 0.05 |
ENSRNOT00000014359
|
Cldn11
|
claudin 11 |
| chr20_+_44680449 | 0.05 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chrX_-_72370044 | 0.05 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr10_+_54246250 | 0.05 |
ENSRNOT00000004880
|
Rcvrn
|
recoverin |
| chr4_-_157263890 | 0.05 |
ENSRNOT00000065416
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr9_-_71778188 | 0.05 |
ENSRNOT00000064562
ENSRNOT00000050886 |
Crygd
|
crystallin, gamma D |
| chr12_-_17904254 | 0.05 |
ENSRNOT00000060192
|
LOC680273
|
similar to Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead-related transcription factor 7) (FREAC-7) |
| chr9_+_41045363 | 0.05 |
ENSRNOT00000066284
|
Cfc1
|
cripto, FRL-1, cryptic family 1 |
| chr12_+_13323547 | 0.05 |
ENSRNOT00000074138
|
Zfp853
|
zinc finger protein 853 |
| chr6_-_138093643 | 0.05 |
ENSRNOT00000045874
|
Igh-6
|
immunoglobulin heavy chain 6 |
| chr7_-_96464049 | 0.05 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr1_-_80689171 | 0.05 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr1_-_80221417 | 0.05 |
ENSRNOT00000072149
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr12_+_7454884 | 0.05 |
ENSRNOT00000077328
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr19_+_46761570 | 0.05 |
ENSRNOT00000058779
|
Wwox
|
WW domain-containing oxidoreductase |
| chr10_-_105628091 | 0.05 |
ENSRNOT00000016067
|
Cygb
|
cytoglobin |
| chrX_-_63809861 | 0.05 |
ENSRNOT00000009870
|
Maged1
|
MAGE family member D1 |
| chr4_-_170932618 | 0.05 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr1_+_72636959 | 0.04 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr11_+_87204175 | 0.04 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr2_+_225645568 | 0.04 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr10_-_90265017 | 0.04 |
ENSRNOT00000064283
ENSRNOT00000048418 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr17_+_82066152 | 0.04 |
ENSRNOT00000083034
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr19_+_25946979 | 0.04 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr13_-_35511020 | 0.04 |
ENSRNOT00000003413
|
Ralb
|
RAS like proto-oncogene B |
| chr7_-_140770647 | 0.04 |
ENSRNOT00000081784
|
C1ql4
|
complement C1q like 4 |
| chr1_+_222519615 | 0.04 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr1_+_82480195 | 0.04 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr1_-_170397191 | 0.04 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr11_+_81796891 | 0.04 |
ENSRNOT00000058402
|
Crygs
|
crystallin, gamma S |
| chr3_+_2642531 | 0.04 |
ENSRNOT00000081798
|
Fut7
|
fucosyltransferase 7 |
| chr5_+_126164674 | 0.04 |
ENSRNOT00000009402
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr1_-_84986581 | 0.04 |
ENSRNOT00000025819
|
Psmc4
|
proteasome 26S subunit, ATPase 4 |
| chr18_+_29110242 | 0.04 |
ENSRNOT00000025756
|
Pura
|
purine rich element binding protein A |
| chr1_-_80615525 | 0.04 |
ENSRNOT00000091574
|
Apoe
|
apolipoprotein E |
| chr1_-_80615704 | 0.04 |
ENSRNOT00000041891
|
Apoe
|
apolipoprotein E |
| chr10_-_88842233 | 0.04 |
ENSRNOT00000026760
|
Stat3
|
signal transducer and activator of transcription 3 |
| chr10_+_105156632 | 0.04 |
ENSRNOT00000086288
|
Galr2
|
galanin receptor 2 |
| chr9_-_81565416 | 0.04 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr12_+_23752844 | 0.04 |
ENSRNOT00000001953
|
Ssc4d
|
scavenger receptor cysteine rich family member with 4 domains |
| chrX_-_15553720 | 0.04 |
ENSRNOT00000012646
|
Gripap1
|
GRIP1 associated protein 1 |
| chr8_+_118525682 | 0.04 |
ENSRNOT00000028288
|
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr10_+_59566223 | 0.03 |
ENSRNOT00000024068
ENSRNOT00000088245 |
P2rx1
|
purinergic receptor P2X 1 |
| chr1_-_212548730 | 0.03 |
ENSRNOT00000089729
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr10_+_91217079 | 0.03 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr4_-_117631716 | 0.03 |
ENSRNOT00000057475
|
LOC103690006
|
RNA/RNP complex-1-interacting phosphatase |
| chr7_-_11777503 | 0.03 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr5_-_147953093 | 0.03 |
ENSRNOT00000075270
|
Khdrbs1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr7_-_58286770 | 0.03 |
ENSRNOT00000005258
|
Rab21
|
RAB21, member RAS oncogene family |
| chr9_-_16611899 | 0.03 |
ENSRNOT00000089168
|
Mea1
|
male-enhanced antigen 1 |
| chr5_+_172273459 | 0.03 |
ENSRNOT00000017957
|
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr12_+_47992091 | 0.03 |
ENSRNOT00000073019
|
Kctd10
|
potassium channel tetramerization domain containing 10 |
| chr7_-_119441487 | 0.03 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr11_+_73738433 | 0.03 |
ENSRNOT00000002353
|
Tmem44
|
transmembrane protein 44 |
| chr16_+_7212488 | 0.03 |
ENSRNOT00000024840
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr7_-_12741296 | 0.03 |
ENSRNOT00000060648
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr10_+_105156822 | 0.03 |
ENSRNOT00000012523
|
Galr2
|
galanin receptor 2 |
| chr1_-_80580662 | 0.03 |
ENSRNOT00000024779
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr19_-_26141111 | 0.03 |
ENSRNOT00000067518
|
Asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr5_-_58476251 | 0.03 |
ENSRNOT00000013151
|
Stoml2
|
stomatin like 2 |
| chrX_-_112328642 | 0.03 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr20_-_45260119 | 0.03 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr1_+_226687258 | 0.03 |
ENSRNOT00000079679
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr3_-_151224123 | 0.03 |
ENSRNOT00000026091
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr12_+_47218969 | 0.03 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr9_-_100930263 | 0.02 |
ENSRNOT00000085029
ENSRNOT00000025644 |
Dtymk
|
deoxythymidylate kinase |
| chr13_-_51076852 | 0.02 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr8_+_67753279 | 0.02 |
ENSRNOT00000009716
|
Calml4
|
calmodulin-like 4 |
| chr19_+_60017746 | 0.02 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr11_+_75905443 | 0.02 |
ENSRNOT00000002650
|
Fgf12
|
fibroblast growth factor 12 |
| chr8_-_111965889 | 0.02 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr18_+_58270410 | 0.02 |
ENSRNOT00000067554
|
Apcdd1
|
APC down-regulated 1 |
| chr7_-_2712723 | 0.02 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr13_-_73638073 | 0.02 |
ENSRNOT00000005195
|
Cep350
|
centrosomal protein 350 |
| chr8_-_75995371 | 0.02 |
ENSRNOT00000014014
|
Foxb1
|
forkhead box B1 |
| chr2_+_227095487 | 0.02 |
ENSRNOT00000019647
|
LOC691807
|
hypothetical protein LOC691807 |
| chr6_+_28663602 | 0.02 |
ENSRNOT00000005402
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr10_+_65507364 | 0.02 |
ENSRNOT00000016297
|
Sdf2
|
stromal cell derived factor 2 |
| chr10_-_11901765 | 0.02 |
ENSRNOT00000009936
ENSRNOT00000075238 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr1_+_266255797 | 0.02 |
ENSRNOT00000027047
|
Trim8
|
tripartite motif-containing 8 |
| chrX_+_119030419 | 0.02 |
ENSRNOT00000060168
|
Pls3
|
plastin 3 |
| chr9_-_16612136 | 0.02 |
ENSRNOT00000023495
|
Mea1
|
male-enhanced antigen 1 |
| chr2_+_187512164 | 0.02 |
ENSRNOT00000051394
|
Mef2d
|
myocyte enhancer factor 2D |
| chr20_+_5125679 | 0.02 |
ENSRNOT00000060832
|
Bag6
|
BCL2-associated athanogene 6 |
| chr9_-_10734073 | 0.02 |
ENSRNOT00000071199
|
Kdm4b
|
lysine demethylase 4B |
| chr8_+_50563047 | 0.02 |
ENSRNOT00000025149
|
Zfp259
|
zinc finger protein 259 |
| chr7_+_70283710 | 0.02 |
ENSRNOT00000075371
|
Ctdsp2
|
CTD small phosphatase 2 |
| chr1_-_220467159 | 0.02 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr2_-_187863349 | 0.02 |
ENSRNOT00000084455
|
Lmna
|
lamin A/C |
| chr3_-_173953684 | 0.02 |
ENSRNOT00000090468
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr17_-_84614228 | 0.02 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chr5_-_57896475 | 0.02 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr4_+_61924013 | 0.02 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr12_+_28212333 | 0.02 |
ENSRNOT00000001182
|
Auts2
|
autism susceptibility candidate 2 |
| chr5_-_144221263 | 0.02 |
ENSRNOT00000013570
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr20_-_10968432 | 0.02 |
ENSRNOT00000001593
|
Cstb
|
cystatin B |
| chr1_+_221099998 | 0.02 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr1_+_79899155 | 0.02 |
ENSRNOT00000078503
|
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr20_+_5125349 | 0.02 |
ENSRNOT00000085598
ENSRNOT00000001129 |
Bag6
|
BCL2-associated athanogene 6 |
| chr2_+_12102487 | 0.02 |
ENSRNOT00000089209
ENSRNOT00000072155 |
LOC100912538
|
centrin-3-like |
| chr11_+_82466071 | 0.02 |
ENSRNOT00000036958
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr8_-_23084879 | 0.02 |
ENSRNOT00000018874
|
Zfp653
|
zinc finger protein 653 |
| chr10_+_85301875 | 0.02 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr2_+_195719543 | 0.02 |
ENSRNOT00000028324
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr5_+_63781801 | 0.02 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr5_-_40867023 | 0.02 |
ENSRNOT00000011576
|
Manea
|
mannosidase, endo-alpha |
| chr12_+_2534212 | 0.02 |
ENSRNOT00000001399
|
Ctxn1
|
cortexin 1 |
| chr4_+_6931495 | 0.02 |
ENSRNOT00000079770
|
Wdr86
|
WD repeat domain 86 |
| chr3_+_11593655 | 0.02 |
ENSRNOT00000074122
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr20_+_3992496 | 0.02 |
ENSRNOT00000082047
|
Psmb8
|
proteasome subunit beta 8 |
| chr19_-_54245855 | 0.02 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr17_+_70586394 | 0.02 |
ENSRNOT00000025550
|
Rbm17
|
RNA binding motif protein 17 |
| chr5_-_59085676 | 0.02 |
ENSRNOT00000068105
|
Msmp
|
microseminoprotein, prostate associated |
| chrX_+_6791136 | 0.02 |
ENSRNOT00000003984
|
LOC100909913
|
norrin-like |
| chr4_-_7311807 | 0.02 |
ENSRNOT00000012222
ENSRNOT00000089817 |
Abcb8
|
ATP binding cassette subfamily B member 8 |
| chr7_+_11677340 | 0.02 |
ENSRNOT00000026648
|
Timm13
|
translocase of inner mitochondrial membrane 13 |
| chr12_-_43940798 | 0.02 |
ENSRNOT00000001485
|
RGD1562310
|
similar to hypothetical protein FLJ21415 |
| chr9_-_99821954 | 0.01 |
ENSRNOT00000090382
ENSRNOT00000022220 |
Otos
|
otospiralin |
| chr1_-_183630695 | 0.01 |
ENSRNOT00000036330
|
AABR07005506.1
|
|
| chr3_+_72447537 | 0.01 |
ENSRNOT00000091901
|
Ssrp1
|
structure specific recognition protein 1 |
| chr10_+_86669233 | 0.01 |
ENSRNOT00000012340
|
Thra
|
thyroid hormone receptor alpha |
| chr3_-_7632345 | 0.01 |
ENSRNOT00000049889
|
Cfap77
|
cilia and flagella associated protein 77 |
| chr16_+_61758917 | 0.01 |
ENSRNOT00000084858
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr5_-_172307431 | 0.01 |
ENSRNOT00000018453
|
Fam213b
|
family with sequence similarity 213, member B |
| chr19_+_53055745 | 0.01 |
ENSRNOT00000074430
|
Foxl1
|
forkhead box L1 |
| chr12_-_21989339 | 0.01 |
ENSRNOT00000074735
|
Ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr3_+_5624506 | 0.01 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr3_+_8450612 | 0.01 |
ENSRNOT00000040457
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr1_+_144070754 | 0.01 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chrX_+_15610230 | 0.01 |
ENSRNOT00000012880
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr13_+_52889737 | 0.01 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr15_+_108608204 | 0.01 |
ENSRNOT00000018918
|
Clybl
|
citrate lyase beta like |
| chr3_+_14482388 | 0.01 |
ENSRNOT00000025857
|
Gsn
|
gelsolin |
| chr9_+_100830250 | 0.01 |
ENSRNOT00000024526
|
Bok
|
BOK, BCL2 family apoptosis regulator |
| chr7_+_133089166 | 0.01 |
ENSRNOT00000085280
|
AC096792.1
|
|
| chr17_+_6683152 | 0.01 |
ENSRNOT00000061249
|
RGD1311345
|
similar to CG9752-PA |
| chr9_-_61690956 | 0.01 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr18_+_73157973 | 0.01 |
ENSRNOT00000031993
|
Skor2
|
SKI family transcriptional corepressor 2 |
| chr13_-_72744295 | 0.01 |
ENSRNOT00000093366
ENSRNOT00000077157 |
Ier5
|
immediate early response 5 |
| chr1_+_198932870 | 0.01 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr1_+_101502987 | 0.01 |
ENSRNOT00000085005
|
Tulp2
|
tubby-like protein 2 |
| chr19_-_56633633 | 0.01 |
ENSRNOT00000023911
|
Ccsap
|
centriole, cilia and spindle-associated protein |
| chrX_-_105390580 | 0.01 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr19_+_25526751 | 0.01 |
ENSRNOT00000083448
|
Cacna1a
|
calcium voltage-gated channel subunit alpha1 A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli3_Zic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.4 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.4 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.0 | 0.1 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.1 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0071031 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.0 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.0 | GO:1990402 | negative regulation of hyaluronan biosynthetic process(GO:1900126) embryonic liver development(GO:1990402) |
| 0.0 | 0.0 | GO:1900453 | negative regulation of long term synaptic depression(GO:1900453) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.0 | GO:0010157 | epidermal cell fate specification(GO:0009957) response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.0 | GO:0015746 | citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.0 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of RNA export from nucleus(GO:0046833) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.0 | 0.4 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0045505 | nitric-oxide synthase regulator activity(GO:0030235) dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |