Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
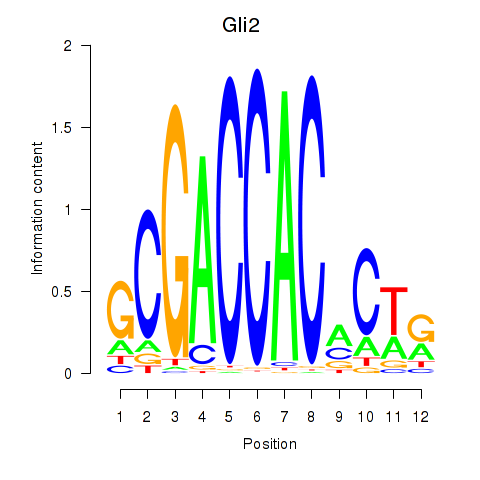
Results for Gli2
Z-value: 0.39
Transcription factors associated with Gli2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli2
|
ENSRNOG00000007261 | GLI family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli2 | rn6_v1_chr13_-_35048444_35048444 | 0.60 | 2.9e-01 | Click! |
Activity profile of Gli2 motif
Sorted Z-values of Gli2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_12658065 | 0.25 |
ENSRNOT00000072951
ENSRNOT00000001679 |
Col6a1
|
collagen type VI alpha 1 chain |
| chr7_-_70552897 | 0.18 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr1_+_141767940 | 0.14 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr3_+_56125924 | 0.14 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr8_+_116332796 | 0.13 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr10_-_27179254 | 0.12 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr8_-_116531784 | 0.12 |
ENSRNOT00000024529
|
Rbm5
|
RNA binding motif protein 5 |
| chrX_-_20070537 | 0.12 |
ENSRNOT00000093602
ENSRNOT00000003397 |
Gnl3l
|
G protein nucleolar 3 like |
| chr6_+_58468155 | 0.12 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr6_+_132702448 | 0.11 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr1_+_215214853 | 0.11 |
ENSRNOT00000071235
ENSRNOT00000080442 |
LOC103690160
|
SH3 and multiple ankyrin repeat domains protein 2-like |
| chr2_-_196526886 | 0.10 |
ENSRNOT00000077325
|
Setdb1
|
SET domain, bifurcated 1 |
| chr8_+_32530806 | 0.10 |
ENSRNOT00000083659
|
Nfrkb
|
nuclear factor related to kappa B binding protein |
| chr10_+_58860940 | 0.10 |
ENSRNOT00000056551
ENSRNOT00000074523 |
XAF1
|
XIAP associated factor-1 |
| chr10_+_104582955 | 0.09 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr12_+_41385241 | 0.09 |
ENSRNOT00000074974
|
Dtx1
|
deltex E3 ubiquitin ligase 1 |
| chr9_-_15700235 | 0.09 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr17_-_90756048 | 0.09 |
ENSRNOT00000085669
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr8_-_116532169 | 0.09 |
ENSRNOT00000085364
|
Rbm5
|
RNA binding motif protein 5 |
| chr14_-_114649173 | 0.09 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr9_+_54558202 | 0.08 |
ENSRNOT00000068433
|
Myo1b
|
myosin Ib |
| chr13_-_83202864 | 0.08 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr10_-_56530842 | 0.08 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chrX_+_53360839 | 0.07 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr5_-_77031120 | 0.07 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr7_-_2353875 | 0.07 |
ENSRNOT00000074873
|
LOC100911319
|
zinc finger protein 36, C3H1 type-like 2-like |
| chr4_+_122282279 | 0.07 |
ENSRNOT00000038244
|
Zxdc
|
ZXD family zinc finger C |
| chr14_-_80169431 | 0.07 |
ENSRNOT00000079769
ENSRNOT00000058315 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr17_-_90755852 | 0.07 |
ENSRNOT00000003526
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr18_+_56887354 | 0.07 |
ENSRNOT00000044346
ENSRNOT00000066133 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr15_-_86105273 | 0.07 |
ENSRNOT00000012600
ENSRNOT00000064942 |
Tbc1d4
|
TBC1 domain family, member 4 |
| chr16_+_23668595 | 0.07 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr18_+_56887722 | 0.06 |
ENSRNOT00000078783
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr5_+_135962911 | 0.06 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chrX_+_29157470 | 0.06 |
ENSRNOT00000081986
|
LOC100910245
|
ribose-phosphate pyrophosphokinase 2-like |
| chr12_-_44999074 | 0.06 |
ENSRNOT00000079177
ENSRNOT00000038293 |
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr9_-_111220651 | 0.06 |
ENSRNOT00000016028
|
Gin1
|
gypsy retrotransposon integrase 1 |
| chr7_+_2718700 | 0.06 |
ENSRNOT00000050792
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr12_+_21721837 | 0.05 |
ENSRNOT00000073353
|
Mepce
|
methylphosphate capping enzyme |
| chr9_-_43022998 | 0.05 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr8_-_22821397 | 0.05 |
ENSRNOT00000045488
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr14_+_39661968 | 0.05 |
ENSRNOT00000064779
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr1_-_281756159 | 0.05 |
ENSRNOT00000013170
|
Prlhr
|
prolactin releasing hormone receptor |
| chr3_-_11410732 | 0.05 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr8_+_22050222 | 0.05 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr9_+_111220858 | 0.04 |
ENSRNOT00000076669
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr10_-_13580821 | 0.04 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr5_-_165442048 | 0.04 |
ENSRNOT00000049822
|
Tardbp
|
TAR DNA binding protein |
| chr7_-_11257977 | 0.04 |
ENSRNOT00000027932
|
Tbxa2r
|
thromboxane A2 receptor |
| chrX_+_73390903 | 0.04 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr2_+_125752130 | 0.04 |
ENSRNOT00000038703
|
Fat4
|
FAT atypical cadherin 4 |
| chr1_+_60106034 | 0.04 |
ENSRNOT00000014930
|
AC091336.1
|
|
| chr6_+_79254339 | 0.04 |
ENSRNOT00000073970
|
Sstr1
|
somatostatin receptor 1 |
| chr14_+_60123169 | 0.04 |
ENSRNOT00000006610
|
Sel1l3
|
SEL1L family member 3 |
| chr19_-_25288335 | 0.04 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr2_-_196527127 | 0.04 |
ENSRNOT00000028709
|
Setdb1
|
SET domain, bifurcated 1 |
| chr8_-_22821223 | 0.04 |
ENSRNOT00000014135
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr3_+_61658245 | 0.04 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr10_+_89089646 | 0.04 |
ENSRNOT00000027505
|
Cntnap1
|
contactin associated protein 1 |
| chr3_-_112371664 | 0.04 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chr5_-_144341586 | 0.03 |
ENSRNOT00000014586
|
Adprhl2
|
ADP-ribosylhydrolase like 2 |
| chr7_+_66595742 | 0.03 |
ENSRNOT00000031191
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr14_-_16452890 | 0.03 |
ENSRNOT00000067222
ENSRNOT00000091577 |
Sept11
|
septin 11 |
| chr8_-_103554399 | 0.03 |
ENSRNOT00000079965
ENSRNOT00000089833 |
LOC100909709
|
short transient receptor potential channel 1-like |
| chr10_+_103934797 | 0.03 |
ENSRNOT00000035865
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr20_+_3555135 | 0.03 |
ENSRNOT00000085380
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_-_11263980 | 0.03 |
ENSRNOT00000025086
|
Nup93
|
nucleoporin 93 |
| chr17_+_16333415 | 0.03 |
ENSRNOT00000060550
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr5_-_144996431 | 0.03 |
ENSRNOT00000016992
|
Zmym4
|
zinc finger MYM-type containing 4 |
| chr8_+_49354115 | 0.02 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr3_-_165741967 | 0.02 |
ENSRNOT00000017063
|
Zfp64
|
zinc finger protein 64 |
| chr7_+_120125633 | 0.02 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr1_+_85112834 | 0.02 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr1_+_221099998 | 0.02 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr4_+_168689163 | 0.02 |
ENSRNOT00000049848
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr18_-_28017925 | 0.02 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr10_-_90265017 | 0.02 |
ENSRNOT00000064283
ENSRNOT00000048418 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr3_-_95418679 | 0.02 |
ENSRNOT00000018074
|
Rcn1
|
reticulocalbin 1 |
| chr10_-_88533829 | 0.02 |
ENSRNOT00000080521
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr5_+_171441560 | 0.02 |
ENSRNOT00000019873
|
Wrap73
|
WD repeat containing, antisense to TP73 |
| chr20_+_12773427 | 0.02 |
ENSRNOT00000001695
|
Col6a2
|
collagen type VI alpha 2 chain |
| chr19_+_10519493 | 0.02 |
ENSRNOT00000030229
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr9_+_47386626 | 0.01 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr9_-_11108741 | 0.01 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr3_+_170354141 | 0.01 |
ENSRNOT00000005901
|
Fam210b
|
family with sequence similarity 210, member B |
| chr20_-_2652952 | 0.01 |
ENSRNOT00000078871
|
C4a
|
complement component 4A (Rodgers blood group) |
| chr6_+_12253788 | 0.01 |
ENSRNOT00000061675
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr2_+_18937458 | 0.01 |
ENSRNOT00000022377
|
Tmem167a
|
transmembrane protein 167A |
| chr14_-_43072843 | 0.01 |
ENSRNOT00000064263
|
Limch1
|
LIM and calponin homology domains 1 |
| chr7_-_11443357 | 0.01 |
ENSRNOT00000082126
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr3_+_172719432 | 0.01 |
ENSRNOT00000033863
|
Zfp831
|
zinc finger protein 831 |
| chrX_+_20216587 | 0.01 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chrX_-_14642424 | 0.01 |
ENSRNOT00000004808
|
Dynlt3
|
dynein light chain Tctex-type 3 |
| chr5_-_144274981 | 0.01 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr1_-_59347472 | 0.01 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr17_-_27452314 | 0.01 |
ENSRNOT00000018936
|
Riok1
|
RIO kinase 1 |
| chr4_+_168976859 | 0.01 |
ENSRNOT00000068663
|
Fam234b
|
family with sequence similarity 234, member B |
| chr20_+_5933303 | 0.01 |
ENSRNOT00000000617
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr3_-_112385528 | 0.01 |
ENSRNOT00000049756
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr18_+_83471342 | 0.01 |
ENSRNOT00000019384
|
Neto1
|
neuropilin and tolloid like 1 |
| chr5_+_67708676 | 0.01 |
ENSRNOT00000075061
|
AABR07048211.1
|
|
| chr4_+_6559545 | 0.01 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr3_+_112371677 | 0.01 |
ENSRNOT00000013316
|
LOC100912076
|
HAUS augmin-like complex subunit 2-like |
| chr6_+_7900972 | 0.01 |
ENSRNOT00000006953
ENSRNOT00000068030 |
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr8_-_107380933 | 0.01 |
ENSRNOT00000022179
|
Pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr20_-_4401610 | 0.01 |
ENSRNOT00000091468
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr1_+_68436917 | 0.00 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr10_+_57462447 | 0.00 |
ENSRNOT00000041443
|
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr12_-_17972737 | 0.00 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr5_-_72000318 | 0.00 |
ENSRNOT00000029187
|
AABR07048308.1
|
|
| chr9_-_100624638 | 0.00 |
ENSRNOT00000051155
|
Hdlbp
|
high density lipoprotein binding protein |
| chr10_+_107502695 | 0.00 |
ENSRNOT00000038088
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr1_+_221043119 | 0.00 |
ENSRNOT00000028185
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr9_-_84101172 | 0.00 |
ENSRNOT00000018652
|
Pax3
|
paired box 3 |
| chr14_+_89253373 | 0.00 |
ENSRNOT00000035922
|
LOC688553
|
hypothetical protein LOC688553 |
| chr7_+_80351774 | 0.00 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.0 | 0.1 | GO:2000537 | immunoglobulin production in mucosal tissue(GO:0002426) T-helper 1 cell activation(GO:0035711) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) regulation of thymocyte migration(GO:2000410) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0021627 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0090073 | negative regulation of protein sumoylation(GO:0033234) positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0042048 | olfactory behavior(GO:0042048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.0 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |