Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
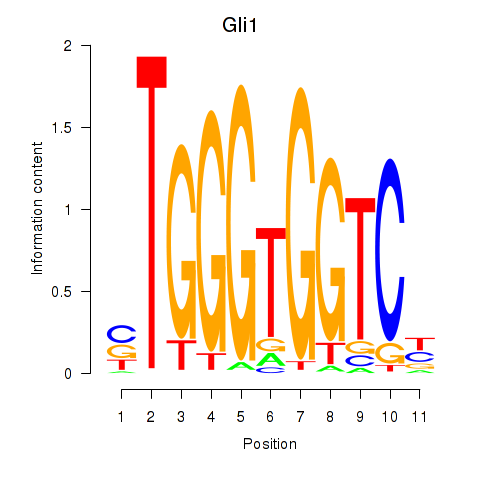
Results for Gli1
Z-value: 0.27
Transcription factors associated with Gli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli1
|
ENSRNOG00000025120 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli1 | rn6_v1_chr7_-_70630338_70630338 | -0.35 | 5.6e-01 | Click! |
Activity profile of Gli1 motif
Sorted Z-values of Gli1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_216080287 | 0.16 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr10_-_90312386 | 0.12 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr6_-_27190126 | 0.11 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr6_+_137997335 | 0.08 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr16_-_82439441 | 0.06 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr1_-_80221417 | 0.06 |
ENSRNOT00000072149
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr3_+_154043873 | 0.06 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr12_+_47074200 | 0.05 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr18_-_28454756 | 0.05 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr4_+_24617008 | 0.05 |
ENSRNOT00000086607
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr1_-_80689171 | 0.04 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr4_+_55715744 | 0.04 |
ENSRNOT00000010429
|
Arf5
|
ADP-ribosylation factor 5 |
| chr2_-_46544457 | 0.04 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr7_-_12326392 | 0.04 |
ENSRNOT00000039728
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chr19_-_37938857 | 0.04 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr3_+_148579920 | 0.04 |
ENSRNOT00000012432
|
Hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr5_+_131380297 | 0.04 |
ENSRNOT00000010548
|
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr5_-_17061361 | 0.04 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr1_-_84812486 | 0.04 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr5_-_60871705 | 0.04 |
ENSRNOT00000016582
|
Exosc3
|
exosome component 3 |
| chr10_+_38889370 | 0.03 |
ENSRNOT00000091236
|
Sept8
|
septin 8 |
| chr5_+_63781801 | 0.03 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr7_-_2712723 | 0.03 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr4_-_170932618 | 0.03 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr19_+_46761570 | 0.03 |
ENSRNOT00000058779
|
Wwox
|
WW domain-containing oxidoreductase |
| chr7_-_96464049 | 0.02 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr7_+_121480723 | 0.02 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr5_+_172273459 | 0.02 |
ENSRNOT00000017957
|
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr10_+_86860685 | 0.02 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr10_+_59566223 | 0.02 |
ENSRNOT00000024068
ENSRNOT00000088245 |
P2rx1
|
purinergic receptor P2X 1 |
| chr4_-_117631716 | 0.02 |
ENSRNOT00000057475
|
LOC103690006
|
RNA/RNP complex-1-interacting phosphatase |
| chr8_-_55177818 | 0.02 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr1_+_207508414 | 0.02 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr18_+_58270410 | 0.02 |
ENSRNOT00000067554
|
Apcdd1
|
APC down-regulated 1 |
| chr10_+_65448950 | 0.02 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr16_+_7212488 | 0.02 |
ENSRNOT00000024840
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr3_-_11102515 | 0.02 |
ENSRNOT00000035580
|
Fam78a
|
family with sequence similarity 78, member A |
| chr18_+_57011575 | 0.02 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chrX_-_72370044 | 0.02 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr16_+_54332660 | 0.02 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr1_+_84304228 | 0.02 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr5_+_135962911 | 0.02 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chr8_+_65686648 | 0.02 |
ENSRNOT00000017233
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr18_-_19275273 | 0.02 |
ENSRNOT00000041707
|
LOC102547344
|
lateral signaling target protein 2 homolog |
| chr11_+_47061354 | 0.02 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr2_+_225645568 | 0.02 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr1_-_261986759 | 0.02 |
ENSRNOT00000021257
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr5_+_126164674 | 0.02 |
ENSRNOT00000009402
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr17_-_84614228 | 0.02 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chr3_+_152752091 | 0.01 |
ENSRNOT00000037177
|
Dlgap4
|
DLG associated protein 4 |
| chrX_-_115426083 | 0.01 |
ENSRNOT00000014756
|
AABR07040947.1
|
|
| chr3_-_148312791 | 0.01 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr17_-_1093873 | 0.01 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr10_-_108321644 | 0.01 |
ENSRNOT00000075171
|
Tbc1d16
|
TBC1 domain family, member 16 |
| chr2_+_198040536 | 0.01 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr3_-_147481073 | 0.01 |
ENSRNOT00000084722
|
Fam110a
|
family with sequence similarity 110, member A |
| chr1_+_192025710 | 0.01 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr10_-_88122233 | 0.01 |
ENSRNOT00000083895
ENSRNOT00000005285 |
Krt14
|
keratin 14 |
| chr7_+_14643704 | 0.01 |
ENSRNOT00000044642
|
AABR07055875.1
|
|
| chr4_+_61924013 | 0.01 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chrX_+_15049462 | 0.01 |
ENSRNOT00000007015
|
Ebp
|
emopamil binding protein (sterol isomerase) |
| chr5_+_140923914 | 0.01 |
ENSRNOT00000020929
|
Heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr9_-_10734073 | 0.01 |
ENSRNOT00000071199
|
Kdm4b
|
lysine demethylase 4B |
| chr2_+_12102487 | 0.01 |
ENSRNOT00000089209
ENSRNOT00000072155 |
LOC100912538
|
centrin-3-like |
| chr10_-_13580821 | 0.01 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr1_-_210739600 | 0.01 |
ENSRNOT00000031734
|
Tcerg1l
|
transcription elongation regulator 1-like |
| chr1_+_144070754 | 0.01 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr10_+_86669233 | 0.01 |
ENSRNOT00000012340
|
Thra
|
thyroid hormone receptor alpha |
| chr5_+_126698582 | 0.01 |
ENSRNOT00000012427
|
Cdcp2
|
CUB domain containing protein 2 |
| chr1_-_220467159 | 0.01 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr1_+_113034227 | 0.01 |
ENSRNOT00000081831
ENSRNOT00000077877 ENSRNOT00000077594 |
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr20_-_10968432 | 0.01 |
ENSRNOT00000001593
|
Cstb
|
cystatin B |
| chr15_+_34155504 | 0.01 |
ENSRNOT00000024782
|
Dhrs4
|
dehydrogenase/reductase 4 |
| chr5_+_133620706 | 0.01 |
ENSRNOT00000053202
|
Gm12830
|
predicted gene 12830 |
| chr1_-_170397191 | 0.01 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr5_-_6186329 | 0.01 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr1_+_72636959 | 0.01 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr1_+_192025357 | 0.00 |
ENSRNOT00000025072
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr18_+_79406381 | 0.00 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr10_+_20320878 | 0.00 |
ENSRNOT00000009714
|
Slit3
|
slit guidance ligand 3 |
| chr8_-_23084879 | 0.00 |
ENSRNOT00000018874
|
Zfp653
|
zinc finger protein 653 |
| chr5_-_65073012 | 0.00 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr7_+_133089166 | 0.00 |
ENSRNOT00000085280
|
AC096792.1
|
|
| chr16_+_61758917 | 0.00 |
ENSRNOT00000084858
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr8_-_59239954 | 0.00 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr1_-_91855983 | 0.00 |
ENSRNOT00000017172
|
Rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr2_+_248715281 | 0.00 |
ENSRNOT00000089648
|
Gtf2b
|
general transcription factor IIB |
| chr19_+_37330930 | 0.00 |
ENSRNOT00000022439
|
Plekhg4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chr19_+_53055745 | 0.00 |
ENSRNOT00000074430
|
Foxl1
|
forkhead box L1 |
| chr7_+_141326950 | 0.00 |
ENSRNOT00000084075
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr6_+_96871625 | 0.00 |
ENSRNOT00000012361
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr3_+_8450612 | 0.00 |
ENSRNOT00000040457
|
Odf2
|
outer dense fiber of sperm tails 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.0 | GO:0071031 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.0 | GO:0051140 | positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |