Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
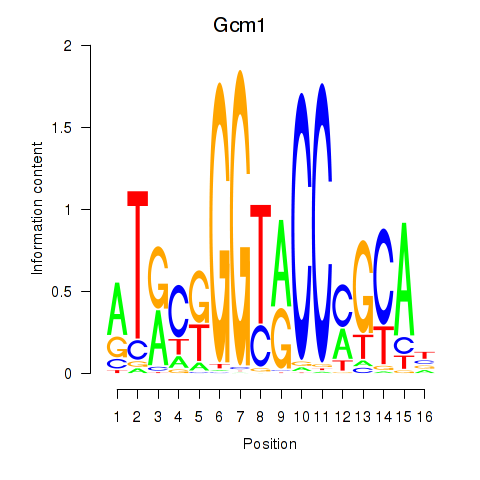
Results for Gcm1
Z-value: 0.39
Transcription factors associated with Gcm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm1
|
ENSRNOG00000007932 | glial cells missing homolog 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gcm1 | rn6_v1_chr8_+_85355766_85355766 | -0.62 | 2.6e-01 | Click! |
Activity profile of Gcm1 motif
Sorted Z-values of Gcm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_115910522 | 0.19 |
ENSRNOT00000076998
ENSRNOT00000067442 |
Arc
|
activity-regulated cytoskeleton-associated protein |
| chr14_+_104191517 | 0.17 |
ENSRNOT00000006573
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_12424367 | 0.14 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr9_+_118586179 | 0.13 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr10_+_61685645 | 0.11 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr3_+_156642464 | 0.11 |
ENSRNOT00000085354
|
Top1
|
topoisomerase (DNA) I |
| chr1_-_125967756 | 0.11 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chrX_+_62363757 | 0.10 |
ENSRNOT00000091240
|
Arx
|
aristaless related homeobox |
| chr3_+_108944141 | 0.10 |
ENSRNOT00000034950
|
Fam98b
|
family with sequence similarity 98, member B |
| chr6_+_135610743 | 0.09 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chrX_+_62363953 | 0.09 |
ENSRNOT00000083362
|
Arx
|
aristaless related homeobox |
| chr13_+_75105615 | 0.09 |
ENSRNOT00000076653
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr7_-_12831675 | 0.09 |
ENSRNOT00000011936
|
Fgf22
|
fibroblast growth factor 22 |
| chrX_+_55331389 | 0.09 |
ENSRNOT00000075300
|
AABR07038567.1
|
|
| chr6_-_49324450 | 0.09 |
ENSRNOT00000066904
|
Sntg2
|
syntrophin, gamma 2 |
| chr8_-_116361343 | 0.08 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr7_-_122247139 | 0.08 |
ENSRNOT00000064515
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chrX_+_68782872 | 0.07 |
ENSRNOT00000075995
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr10_+_102136283 | 0.07 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr15_+_34187223 | 0.07 |
ENSRNOT00000024978
|
Cpne6
|
copine 6 |
| chr14_-_78377825 | 0.06 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr3_+_33440191 | 0.06 |
ENSRNOT00000034632
ENSRNOT00000092907 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr8_+_96551245 | 0.06 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr11_-_87158597 | 0.05 |
ENSRNOT00000002517
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr2_-_186480278 | 0.05 |
ENSRNOT00000031743
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chr5_+_139468546 | 0.05 |
ENSRNOT00000013193
|
Slfnl1
|
schlafen-like 1 |
| chr6_-_147172022 | 0.05 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr3_-_122947075 | 0.05 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr1_-_101131413 | 0.05 |
ENSRNOT00000093729
|
Flt3lg
|
fms-related tyrosine kinase 3 ligand |
| chr2_+_127459012 | 0.04 |
ENSRNOT00000093685
ENSRNOT00000072675 |
Intu
|
inturned planar cell polarity protein |
| chr9_+_49479023 | 0.04 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr5_-_151397603 | 0.04 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr14_-_20953095 | 0.04 |
ENSRNOT00000004440
|
Dck
|
deoxycytidine kinase |
| chr8_+_7128656 | 0.04 |
ENSRNOT00000038313
|
Pgr
|
progesterone receptor |
| chr9_+_47386626 | 0.04 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr1_+_266530477 | 0.03 |
ENSRNOT00000054699
|
Cnnm2
|
cyclin and CBS domain divalent metal cation transport mediator 2 |
| chr13_-_111849653 | 0.03 |
ENSRNOT00000006395
|
Diexf
|
digestive organ expansion factor homolog (zebrafish) |
| chr11_-_86276430 | 0.03 |
ENSRNOT00000075164
|
Hira
|
histone cell cycle regulator |
| chr6_+_18821815 | 0.03 |
ENSRNOT00000062057
|
Mtm1
|
myotubularin 1 |
| chr1_-_101131012 | 0.03 |
ENSRNOT00000082283
ENSRNOT00000093498 ENSRNOT00000093559 |
Flt3lg
Rpl13a
|
fms-related tyrosine kinase 3 ligand ribosomal protein L13A |
| chr2_+_57206613 | 0.03 |
ENSRNOT00000082694
ENSRNOT00000046069 |
Nup155
|
nucleoporin 155 |
| chr14_-_104523297 | 0.03 |
ENSRNOT00000039523
|
Cep68
|
centrosomal protein 68 |
| chr3_+_122308025 | 0.03 |
ENSRNOT00000008071
|
Stk35
|
serine/threonine kinase 35 |
| chrX_+_18998532 | 0.02 |
ENSRNOT00000004213
|
AABR07037343.1
|
|
| chr15_+_3996328 | 0.02 |
ENSRNOT00000088252
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr1_-_220165678 | 0.02 |
ENSRNOT00000026915
|
Bbs1
|
Bardet-Biedl syndrome 1 |
| chr1_+_220423426 | 0.02 |
ENSRNOT00000072647
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr7_+_117963740 | 0.02 |
ENSRNOT00000075405
|
LOC108348189
|
COMM domain-containing protein 5 |
| chr2_+_243502073 | 0.02 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_7311807 | 0.02 |
ENSRNOT00000012222
ENSRNOT00000089817 |
Abcb8
|
ATP binding cassette subfamily B member 8 |
| chr15_+_3996048 | 0.02 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr10_+_1464765 | 0.01 |
ENSRNOT00000004217
|
Bfar
|
bifunctional apoptosis regulator |
| chr5_-_141033511 | 0.01 |
ENSRNOT00000045922
|
Bmp8a
|
bone morphogenetic protein 8a |
| chr4_-_113764532 | 0.01 |
ENSRNOT00000009269
|
Sema4f
|
ssemaphorin 4F |
| chr1_-_201906286 | 0.01 |
ENSRNOT00000064511
ENSRNOT00000036625 |
RGD1560958
|
similar to RIKEN cDNA 1700063I17 |
| chr18_+_3958432 | 0.01 |
ENSRNOT00000073509
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr19_+_37127508 | 0.01 |
ENSRNOT00000019656
|
Cbfb
|
core-binding factor, beta subunit |
| chr6_-_50846965 | 0.01 |
ENSRNOT00000087300
|
Slc26a4
|
solute carrier family 26 member 4 |
| chr20_+_50394650 | 0.01 |
ENSRNOT00000076010
ENSRNOT00000077065 ENSRNOT00000073259 |
Popdc3
|
popeye domain containing 3 |
| chr1_+_53360157 | 0.01 |
ENSRNOT00000017809
|
Rps6ka2
|
ribosomal protein S6 kinase A2 |
| chr5_+_157165341 | 0.01 |
ENSRNOT00000087072
|
Pla2g2c
|
phospholipase A2, group IIC |
| chr1_-_259089632 | 0.01 |
ENSRNOT00000020940
ENSRNOT00000091297 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_-_153252021 | 0.01 |
ENSRNOT00000023221
|
Tmem50a
|
transmembrane protein 50A |
| chr1_-_259674425 | 0.01 |
ENSRNOT00000091116
|
LOC108348083
|
delta-1-pyrroline-5-carboxylate synthase |
| chr10_+_105861743 | 0.01 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr16_+_81089292 | 0.01 |
ENSRNOT00000087192
ENSRNOT00000026150 |
Tfdp1
|
transcription factor Dp-1 |
| chr7_-_117267402 | 0.01 |
ENSRNOT00000088945
|
Plec
|
plectin |
| chr12_-_21450204 | 0.00 |
ENSRNOT00000065866
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr19_-_17346808 | 0.00 |
ENSRNOT00000017361
|
Rbl2
|
RB transcriptional corepressor like 2 |
| chr9_-_85528860 | 0.00 |
ENSRNOT00000063834
ENSRNOT00000088963 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr6_-_147172813 | 0.00 |
ENSRNOT00000066545
|
Itgb8
|
integrin subunit beta 8 |
| chr14_-_3389943 | 0.00 |
ENSRNOT00000037101
|
Ephx4
|
epoxide hydrolase 4 |
| chr2_-_123323170 | 0.00 |
ENSRNOT00000021453
|
Bbs7
|
Bardet-Biedl syndrome 7 |
| chr1_+_274245184 | 0.00 |
ENSRNOT00000018889
|
Dusp5
|
dual specificity phosphatase 5 |
| chr10_+_105796680 | 0.00 |
ENSRNOT00000000264
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gcm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.2 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |