Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
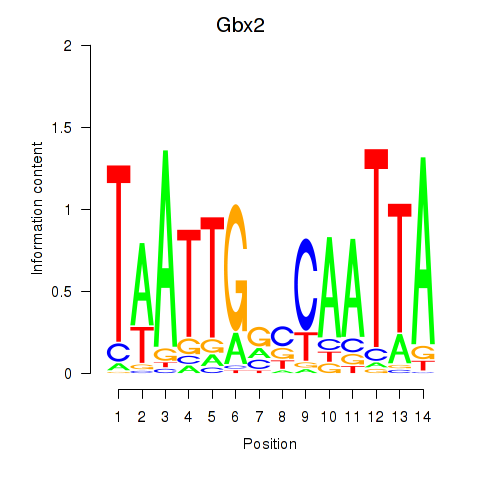
Results for Gbx2
Z-value: 0.20
Transcription factors associated with Gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx2
|
ENSRNOG00000019495 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gbx2 | rn6_v1_chr9_-_97065817_97065817 | -0.80 | 1.1e-01 | Click! |
Activity profile of Gbx2 motif
Sorted Z-values of Gbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_11497531 | 0.07 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_+_11912543 | 0.06 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr12_-_17322608 | 0.06 |
ENSRNOT00000033038
|
LOC102546864
|
uncharacterized LOC102546864 |
| chr9_+_73378057 | 0.05 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr13_+_80464348 | 0.05 |
ENSRNOT00000076324
|
Vamp4
|
vesicle-associated membrane protein 4 |
| chr2_+_86996497 | 0.05 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr13_-_83457888 | 0.04 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr2_+_86996798 | 0.04 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr8_-_45137893 | 0.04 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr6_-_108660063 | 0.04 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr2_+_72006099 | 0.04 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr2_+_196608496 | 0.04 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr3_-_90751055 | 0.04 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr16_+_22361998 | 0.03 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr1_+_220071811 | 0.03 |
ENSRNOT00000090642
|
AC126581.1
|
|
| chr1_-_78180216 | 0.03 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr12_-_46493203 | 0.03 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr3_+_5624506 | 0.03 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr3_-_138683318 | 0.03 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr11_+_38727048 | 0.02 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr2_+_220432037 | 0.02 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr3_+_48106099 | 0.02 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chrX_-_25628272 | 0.02 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr16_-_10802512 | 0.02 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr2_-_199971965 | 0.02 |
ENSRNOT00000087026
|
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr17_-_80860195 | 0.02 |
ENSRNOT00000038651
|
Trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chrX_+_84064427 | 0.02 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr7_-_14364178 | 0.02 |
ENSRNOT00000090673
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chr2_+_2903492 | 0.02 |
ENSRNOT00000062017
|
Ttc37
|
tetratricopeptide repeat domain 37 |
| chr18_+_52423883 | 0.01 |
ENSRNOT00000022017
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr1_+_137014272 | 0.01 |
ENSRNOT00000014802
|
Akap13
|
A-kinase anchoring protein 13 |
| chr1_-_129776276 | 0.01 |
ENSRNOT00000051402
|
Arrdc4
|
arrestin domain containing 4 |
| chrX_+_78042859 | 0.01 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr11_+_40509078 | 0.01 |
ENSRNOT00000023108
|
AABR07033851.1
|
|
| chr10_+_95770154 | 0.01 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr5_+_135574172 | 0.01 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr12_-_48663143 | 0.01 |
ENSRNOT00000082809
|
Ficd
|
FIC domain containing |
| chr1_+_228395558 | 0.01 |
ENSRNOT00000065411
|
Osbp
|
oxysterol binding protein |
| chrX_+_14578264 | 0.01 |
ENSRNOT00000038994
|
Cybb
|
cytochrome b-245 beta chain |
| chr6_-_136436620 | 0.01 |
ENSRNOT00000067118
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr5_+_163773509 | 0.01 |
ENSRNOT00000072167
|
LOC103692519
|
60S ribosomal protein L9 pseudogene |
| chr5_+_163612518 | 0.01 |
ENSRNOT00000071103
|
LOC103692519
|
60S ribosomal protein L9 pseudogene |
| chr2_-_189333015 | 0.01 |
ENSRNOT00000076424
ENSRNOT00000076952 |
Hax1
|
HCLS1 associated protein X-1 |
| chr7_-_107223047 | 0.01 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr6_-_127632265 | 0.01 |
ENSRNOT00000084157
|
Serpina1
|
serpin family A member 1 |
| chrX_+_131381134 | 0.01 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr2_+_54466280 | 0.01 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr14_-_106248539 | 0.01 |
ENSRNOT00000067668
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr10_-_87521514 | 0.01 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr5_-_9035811 | 0.01 |
ENSRNOT00000082023
|
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr20_+_3155652 | 0.01 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr16_+_6048004 | 0.00 |
ENSRNOT00000020750
|
Actr8
|
ARP8 actin-related protein 8 homolog |
| chr18_+_16146447 | 0.00 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr1_-_23556241 | 0.00 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr1_+_147713892 | 0.00 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr1_+_79631668 | 0.00 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr8_+_79638696 | 0.00 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr1_+_265157379 | 0.00 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr14_-_18704059 | 0.00 |
ENSRNOT00000081455
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr4_-_169036950 | 0.00 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr3_-_14386118 | 0.00 |
ENSRNOT00000025649
|
Rab14
|
RAB14, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |