Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
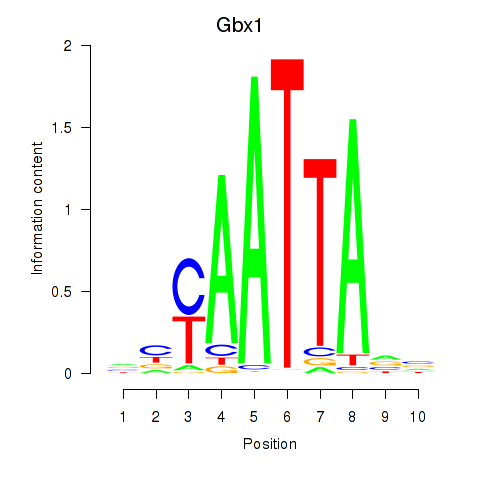
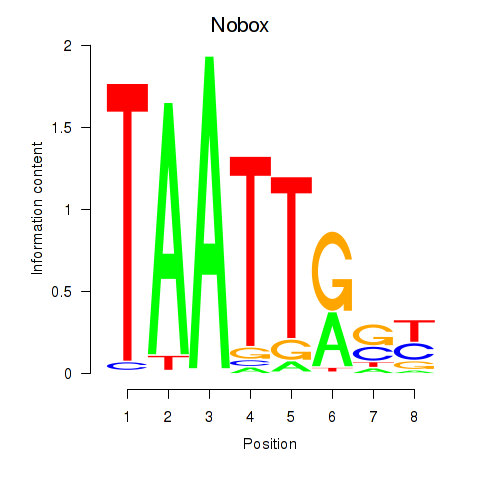
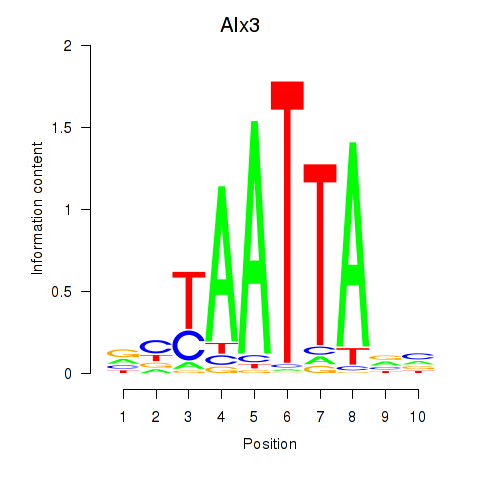
Results for Gbx1_Nobox_Alx3
Z-value: 0.22
Transcription factors associated with Gbx1_Nobox_Alx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx1
|
ENSRNOG00000013369 | gastrulation brain homeobox 1 |
|
Nobox
|
ENSRNOG00000025720 | NOBOX oogenesis homeobox |
|
Alx3
|
ENSRNOG00000018290 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nobox | rn6_v1_chr4_-_72756887_72756887 | 0.68 | 2.0e-01 | Click! |
| Gbx1 | rn6_v1_chr4_+_7173961_7173961 | 0.45 | 4.5e-01 | Click! |
| Alx3 | rn6_v1_chr2_+_210381829_210381829 | -0.26 | 6.8e-01 | Click! |
Activity profile of Gbx1_Nobox_Alx3 motif
Sorted Z-values of Gbx1_Nobox_Alx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 0.78 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_-_66417741 | 0.22 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr12_-_45801842 | 0.17 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr12_-_17186679 | 0.16 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr2_+_145174876 | 0.16 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr6_-_115616766 | 0.12 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr19_+_25043680 | 0.11 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr6_-_106971250 | 0.10 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr10_-_88670430 | 0.10 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr6_-_125723732 | 0.08 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr2_-_33025271 | 0.08 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_215033460 | 0.08 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr20_+_3558827 | 0.08 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr18_-_31343797 | 0.07 |
ENSRNOT00000081850
|
Pcdh1
|
protocadherin 1 |
| chr3_+_95715193 | 0.06 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr4_+_158088505 | 0.05 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr15_-_30147793 | 0.05 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr1_+_13595295 | 0.05 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr6_-_111417813 | 0.05 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr1_+_105284753 | 0.05 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chrX_-_124464963 | 0.05 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chrX_+_118197217 | 0.05 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr12_-_35979193 | 0.04 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr1_-_43638161 | 0.04 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr13_+_90533365 | 0.04 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr5_-_12526962 | 0.04 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr10_-_84920886 | 0.04 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr10_-_74679858 | 0.04 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr10_-_87286387 | 0.04 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr17_+_11683862 | 0.04 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr20_+_44680449 | 0.04 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr9_+_117795132 | 0.03 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr4_+_88694583 | 0.03 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr1_-_89539210 | 0.03 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr6_+_58468155 | 0.03 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chrX_+_124321551 | 0.03 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr1_+_217345545 | 0.03 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr6_+_73553210 | 0.03 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chrX_+_6273733 | 0.03 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr2_-_187786700 | 0.03 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr3_-_161917285 | 0.03 |
ENSRNOT00000025150
|
Cdh22
|
cadherin 22 |
| chr5_-_168734296 | 0.03 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr18_-_26656879 | 0.03 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr13_+_34400170 | 0.03 |
ENSRNOT00000061516
ENSRNOT00000061515 ENSRNOT00000061513 ENSRNOT00000084506 ENSRNOT00000086641 |
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr2_+_266315036 | 0.03 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr7_+_136182224 | 0.03 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr7_-_142063212 | 0.03 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr1_+_278557792 | 0.03 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr10_-_91986632 | 0.03 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr6_+_8219385 | 0.03 |
ENSRNOT00000040509
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr1_-_278042312 | 0.03 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chrX_+_74200972 | 0.02 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr16_+_7035241 | 0.02 |
ENSRNOT00000084977
|
Nek4
|
NIMA-related kinase 4 |
| chr15_+_44441856 | 0.02 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr14_+_104250617 | 0.02 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr10_-_93675991 | 0.02 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr4_-_17594598 | 0.02 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr3_+_128828331 | 0.02 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr7_+_2752680 | 0.02 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr4_+_68656928 | 0.02 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr14_-_21128505 | 0.02 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_-_166912524 | 0.02 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr19_+_39229754 | 0.02 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr8_-_78233430 | 0.02 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr3_+_48106099 | 0.02 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr8_+_71914867 | 0.02 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr1_-_7480825 | 0.02 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr16_-_930527 | 0.02 |
ENSRNOT00000014787
|
Spin1
|
spindlin 1 |
| chr3_+_45683993 | 0.02 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_+_195996521 | 0.02 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr4_-_66955732 | 0.02 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr18_+_51523758 | 0.02 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr1_+_185210922 | 0.02 |
ENSRNOT00000055120
|
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr5_-_136965191 | 0.02 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr1_-_48825364 | 0.02 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr4_+_117962319 | 0.02 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr3_-_11452529 | 0.01 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr9_+_71915421 | 0.01 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr16_-_25192675 | 0.01 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr10_+_91217079 | 0.01 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr1_+_140998240 | 0.01 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr1_+_204959174 | 0.01 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr6_-_4520604 | 0.01 |
ENSRNOT00000042230
ENSRNOT00000043870 ENSRNOT00000070918 ENSRNOT00000046246 ENSRNOT00000052367 ENSRNOT00000042251 |
Slc8a1
|
solute carrier family 8 member A1 |
| chr3_-_60765645 | 0.01 |
ENSRNOT00000050513
|
Atf2
|
activating transcription factor 2 |
| chr13_+_31081804 | 0.01 |
ENSRNOT00000041413
|
Cdh7
|
cadherin 7 |
| chr5_-_145423172 | 0.01 |
ENSRNOT00000081919
|
Gjb5
|
gap junction protein, beta 5 |
| chr11_-_62067655 | 0.01 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_-_87826421 | 0.01 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr5_+_135574172 | 0.01 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr12_-_41485122 | 0.01 |
ENSRNOT00000001859
|
Ddx54
|
DEAD-box helicase 54 |
| chr3_+_56125924 | 0.01 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr19_-_58735173 | 0.01 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr7_-_122329443 | 0.01 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr2_-_188559882 | 0.01 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr1_-_101819478 | 0.01 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr16_+_7035068 | 0.01 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chr18_+_79773608 | 0.01 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr3_+_148654668 | 0.01 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr4_+_22898527 | 0.01 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr3_-_57607683 | 0.01 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr1_+_28454966 | 0.01 |
ENSRNOT00000078841
ENSRNOT00000030327 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr3_+_48096954 | 0.01 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr8_-_48597867 | 0.01 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chr16_-_10802512 | 0.01 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr2_+_80269661 | 0.01 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr16_-_74264142 | 0.01 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr7_+_78558701 | 0.01 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_70242874 | 0.01 |
ENSRNOT00000010924
|
Fndc8
|
fibronectin type III domain containing 8 |
| chrX_-_104932508 | 0.01 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr13_+_98311827 | 0.01 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_-_227932603 | 0.01 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr14_-_16903242 | 0.01 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr18_+_57011575 | 0.01 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr1_-_23556241 | 0.01 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr2_+_83393282 | 0.01 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr1_-_64147251 | 0.01 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chrX_+_74205842 | 0.01 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr9_-_81566096 | 0.01 |
ENSRNOT00000019497
|
Aamp
|
angio-associated, migratory cell protein |
| chr4_+_61924013 | 0.01 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr3_-_103745236 | 0.01 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr9_+_73378057 | 0.01 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr18_-_16543992 | 0.01 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr16_+_60925093 | 0.01 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr19_+_22699808 | 0.01 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr19_+_45938915 | 0.01 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr3_+_113423693 | 0.01 |
ENSRNOT00000021091
|
Hypk
|
Huntingtin interacting protein K |
| chr2_-_165600748 | 0.01 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr1_+_260093641 | 0.01 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr19_-_17399548 | 0.01 |
ENSRNOT00000075991
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_148845267 | 0.01 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr15_-_95514259 | 0.01 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr10_-_13446135 | 0.01 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr11_-_62451149 | 0.01 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr16_-_9658484 | 0.01 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr2_+_51672722 | 0.01 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr9_-_105693357 | 0.01 |
ENSRNOT00000066968
|
Nudt12
|
nudix hydrolase 12 |
| chr6_+_42854888 | 0.01 |
ENSRNOT00000007259
|
Odc1
|
ornithine decarboxylase 1 |
| chr1_-_128721956 | 0.01 |
ENSRNOT00000076511
ENSRNOT00000050832 |
Synm
|
synemin |
| chr7_-_14364178 | 0.01 |
ENSRNOT00000090673
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chr1_+_167870452 | 0.01 |
ENSRNOT00000025027
|
Olr63
|
olfactory receptor 63 |
| chr16_+_71787966 | 0.01 |
ENSRNOT00000080084
|
Htra4
|
HtrA serine peptidase 4 |
| chr1_-_134871167 | 0.01 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr5_-_7941822 | 0.01 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr9_-_11027506 | 0.01 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr13_+_87986240 | 0.01 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr6_+_72124417 | 0.01 |
ENSRNOT00000040548
|
Scfd1
|
sec1 family domain containing 1 |
| chr7_+_144865608 | 0.01 |
ENSRNOT00000091596
ENSRNOT00000055285 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr7_+_42304534 | 0.01 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr3_+_16846412 | 0.01 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr9_-_85243001 | 0.00 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr14_+_39964588 | 0.00 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr15_-_61648267 | 0.00 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr3_-_37803112 | 0.00 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr10_-_87232723 | 0.00 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr2_+_154604832 | 0.00 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr1_+_225037737 | 0.00 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr2_+_136993208 | 0.00 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr5_+_146656049 | 0.00 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr4_-_100252755 | 0.00 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr14_-_19072677 | 0.00 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr1_-_85517360 | 0.00 |
ENSRNOT00000026114
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr2_+_195651930 | 0.00 |
ENSRNOT00000028299
|
Tdrkh
|
tudor and KH domain containing |
| chrX_-_158978995 | 0.00 |
ENSRNOT00000001179
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr2_+_165076607 | 0.00 |
ENSRNOT00000012831
|
Il12a
|
interleukin 12A |
| chr15_+_31950986 | 0.00 |
ENSRNOT00000080233
|
AABR07017868.4
|
|
| chr8_-_39551700 | 0.00 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr8_+_33239139 | 0.00 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_+_158097843 | 0.00 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr6_+_106496992 | 0.00 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr5_+_133819726 | 0.00 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chr2_+_118910587 | 0.00 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
| chrX_-_62690806 | 0.00 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr9_+_50966766 | 0.00 |
ENSRNOT00000076636
|
Ercc5
|
ERCC excision repair 5, endonuclease |
| chr9_+_49647257 | 0.00 |
ENSRNOT00000021899
|
Mrps9
|
mitochondrial ribosomal protein S9 |
| chr1_-_228753422 | 0.00 |
ENSRNOT00000028626
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr4_+_68608137 | 0.00 |
ENSRNOT00000036398
|
Wee2
|
WEE1 homolog 2 |
| chr10_-_16731898 | 0.00 |
ENSRNOT00000028186
|
Crebrf
|
CREB3 regulatory factor |
| chr16_+_7758996 | 0.00 |
ENSRNOT00000061063
|
Btd
|
biotinidase |
| chr4_-_21920651 | 0.00 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr19_+_38039729 | 0.00 |
ENSRNOT00000089783
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chr1_+_80141630 | 0.00 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr8_+_12994155 | 0.00 |
ENSRNOT00000011855
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr10_-_86004096 | 0.00 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr16_-_73152921 | 0.00 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chr14_+_37116492 | 0.00 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr1_-_228505080 | 0.00 |
ENSRNOT00000028600
|
Olr322
|
olfactory receptor 322 |
| chr1_+_68436593 | 0.00 |
ENSRNOT00000080325
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr4_-_157304653 | 0.00 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr7_+_44009069 | 0.00 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr2_-_123972356 | 0.00 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr6_-_1003905 | 0.00 |
ENSRNOT00000082071
ENSRNOT00000042735 |
Fez2
|
fasciculation and elongation protein zeta 2 |
| chr5_+_36566783 | 0.00 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr11_-_61287914 | 0.00 |
ENSRNOT00000086286
|
Spice1
|
spindle and centriole associated protein 1 |
| chr19_+_6046665 | 0.00 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr2_-_44786963 | 0.00 |
ENSRNOT00000060695
|
Skiv2l2
|
Ski2 like RNA helicase 2 |
| chr8_-_53816447 | 0.00 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr13_+_49005405 | 0.00 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr11_+_31539016 | 0.00 |
ENSRNOT00000072856
|
Ifnar2
|
interferon alpha and beta receptor subunit 2 |
| chr1_-_15374850 | 0.00 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr3_+_140024043 | 0.00 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx1_Nobox_Alx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |