Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
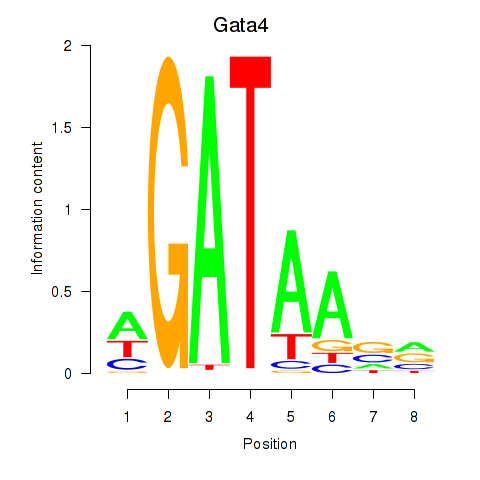
Results for Gata4
Z-value: 0.23
Transcription factors associated with Gata4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata4
|
ENSRNOG00000010708 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata4 | rn6_v1_chr15_-_46432965_46432965 | 0.48 | 4.1e-01 | Click! |
Activity profile of Gata4 motif
Sorted Z-values of Gata4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_50381244 | 0.13 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr17_-_22143324 | 0.10 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr1_+_279798187 | 0.10 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr9_-_82146874 | 0.10 |
ENSRNOT00000024190
|
Fev
|
FEV, ETS transcription factor |
| chr1_+_72882806 | 0.10 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr1_-_261446570 | 0.10 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr1_-_164205222 | 0.06 |
ENSRNOT00000035774
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr10_+_103874383 | 0.05 |
ENSRNOT00000038935
|
Otop2
|
otopetrin 2 |
| chr8_+_50525091 | 0.04 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr15_+_67555835 | 0.04 |
ENSRNOT00000045882
|
Pcdh17
|
protocadherin 17 |
| chr3_+_160852164 | 0.04 |
ENSRNOT00000019127
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr8_-_33661049 | 0.04 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr18_-_16542165 | 0.04 |
ENSRNOT00000079381
|
Slc39a6
|
solute carrier family 39 member 6 |
| chrX_+_112270986 | 0.04 |
ENSRNOT00000091441
ENSRNOT00000082652 |
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr5_+_138685624 | 0.04 |
ENSRNOT00000011867
|
Guca2a
|
guanylate cyclase activator 2A |
| chr1_+_175445088 | 0.04 |
ENSRNOT00000036718
|
Adm
|
adrenomedullin |
| chr13_-_53870428 | 0.03 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr19_-_14945302 | 0.03 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr8_-_117820413 | 0.02 |
ENSRNOT00000075819
|
Tma7
|
translation machinery associated 7 homolog |
| chr4_-_181477281 | 0.02 |
ENSRNOT00000055463
|
Mansc4
|
MANSC domain containing 4 |
| chrX_-_78496847 | 0.02 |
ENSRNOT00000003265
|
Itm2a
|
integral membrane protein 2A |
| chr10_-_15610826 | 0.02 |
ENSRNOT00000027851
|
Hbz
|
hemoglobin subunit zeta |
| chr6_+_50725085 | 0.02 |
ENSRNOT00000009473
|
Slc26a3
|
solute carrier family 26 member 3 |
| chr20_-_2191640 | 0.02 |
ENSRNOT00000001016
|
Trim10
|
tripartite motif-containing 10 |
| chr4_+_119225040 | 0.02 |
ENSRNOT00000012365
|
Bmp10
|
bone morphogenetic protein 10 |
| chrX_-_13601069 | 0.02 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr10_-_87232723 | 0.02 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr15_-_61772516 | 0.02 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr19_-_10450186 | 0.02 |
ENSRNOT00000088048
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_-_60679555 | 0.02 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr3_+_79918969 | 0.02 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr11_-_33003021 | 0.02 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr1_-_226732736 | 0.02 |
ENSRNOT00000072343
|
LOC108348129
|
pepsin F-like |
| chr4_+_163162211 | 0.02 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr6_+_22696397 | 0.02 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chr2_+_116970344 | 0.02 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr11_-_35749464 | 0.02 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr8_+_22047697 | 0.02 |
ENSRNOT00000067741
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr3_-_112985318 | 0.02 |
ENSRNOT00000015556
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr2_+_42726809 | 0.02 |
ENSRNOT00000036860
|
AABR07008097.1
|
|
| chr9_+_95256627 | 0.02 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr7_+_11490852 | 0.02 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr5_+_133864798 | 0.01 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr6_+_137164535 | 0.01 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr5_-_28130803 | 0.01 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr6_+_7961413 | 0.01 |
ENSRNOT00000007638
ENSRNOT00000061871 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr5_+_154077944 | 0.01 |
ENSRNOT00000031512
|
Il22ra1
|
interleukin 22 receptor subunit alpha 1 |
| chr11_-_64952687 | 0.01 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr5_+_14890408 | 0.01 |
ENSRNOT00000033116
|
Sox17
|
SRY box 17 |
| chr2_-_60657712 | 0.01 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr3_+_159936856 | 0.01 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chrX_+_83678339 | 0.01 |
ENSRNOT00000005997
|
Apool
|
apolipoprotein O-like |
| chr2_+_189423559 | 0.01 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr13_+_48513008 | 0.01 |
ENSRNOT00000009742
|
Slc26a9
|
solute carrier family 26 member 9 |
| chr16_+_49266903 | 0.01 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr15_+_33632416 | 0.01 |
ENSRNOT00000068212
|
AC130940.1
|
|
| chr17_-_21677477 | 0.01 |
ENSRNOT00000035448
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr19_-_25166528 | 0.01 |
ENSRNOT00000007775
|
Rln3
|
relaxin 3 |
| chr1_+_17602281 | 0.01 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr10_-_90415070 | 0.01 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr3_+_55910177 | 0.01 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr6_-_26385761 | 0.01 |
ENSRNOT00000073228
|
Gckr
|
glucokinase regulator |
| chr3_-_2453933 | 0.01 |
ENSRNOT00000014060
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr2_-_77784522 | 0.01 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr17_-_84247038 | 0.01 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr17_-_21705773 | 0.01 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_-_259382765 | 0.01 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr8_-_48672732 | 0.01 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr3_-_93216495 | 0.01 |
ENSRNOT00000010580
|
Ehf
|
ets homologous factor |
| chr5_-_28131133 | 0.01 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr10_+_70262361 | 0.01 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_-_192027225 | 0.01 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr6_-_23291568 | 0.01 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr6_-_132797675 | 0.01 |
ENSRNOT00000006128
|
Wars
|
tryptophanyl-tRNA synthetase |
| chr19_-_38344845 | 0.00 |
ENSRNOT00000072666
|
AABR07043748.1
|
|
| chrX_-_104932508 | 0.00 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr11_+_80736576 | 0.00 |
ENSRNOT00000047678
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr16_+_50022998 | 0.00 |
ENSRNOT00000087986
|
Tlr3
|
toll-like receptor 3 |
| chr7_+_60099120 | 0.00 |
ENSRNOT00000007338
|
LOC100911101
|
leucine-rich repeat-containing protein 10-like |
| chr4_-_163261958 | 0.00 |
ENSRNOT00000086390
|
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr20_+_7718282 | 0.00 |
ENSRNOT00000000593
|
Scube3
|
signal peptide, CUB domain and EGF like domain containing 3 |
| chr2_+_212257225 | 0.00 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chrX_+_20351486 | 0.00 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chrX_-_119162518 | 0.00 |
ENSRNOT00000073176
|
LOC103694506
|
ubiquitin-conjugating enzyme E2 W |
| chr1_-_85220237 | 0.00 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr14_-_45165207 | 0.00 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr20_-_27657983 | 0.00 |
ENSRNOT00000057313
|
Fam26e
|
family with sequence similarity 26, member E |
| chr6_-_7961207 | 0.00 |
ENSRNOT00000007174
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr15_-_11812485 | 0.00 |
ENSRNOT00000030991
ENSRNOT00000007795 |
Nek10
|
NIMA-related kinase 10 |
| chr2_+_147496229 | 0.00 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.0 | 0.1 | GO:2000041 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |