Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Gata2_Gata1
Z-value: 0.40
Transcription factors associated with Gata2_Gata1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
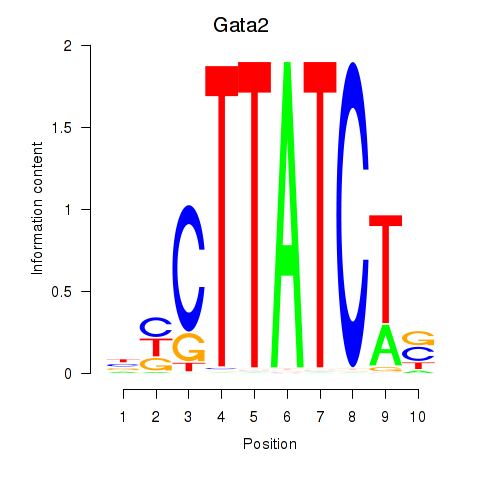
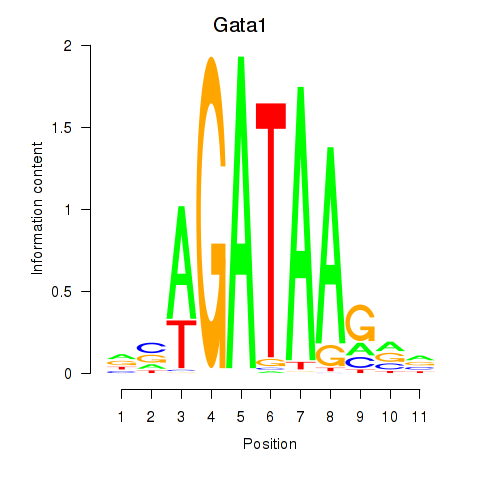
Gata2
|
ENSRNOG00000012347 | GATA binding protein 2 |
|
Gata1
|
ENSRNOG00000025534 | GATA binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata2 | rn6_v1_chr4_+_120133713_120133713 | -0.29 | 6.3e-01 | Click! |
| Gata1 | rn6_v1_chrX_+_15378789_15378789 | 0.21 | 7.3e-01 | Click! |
Activity profile of Gata2_Gata1 motif
Sorted Z-values of Gata2_Gata1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_72882806 | 0.23 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr1_-_261446570 | 0.23 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr2_-_48501436 | 0.20 |
ENSRNOT00000017305
|
Isl1
|
ISL LIM homeobox 1 |
| chr10_+_103874383 | 0.20 |
ENSRNOT00000038935
|
Otop2
|
otopetrin 2 |
| chr17_-_22143324 | 0.20 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr8_-_33661049 | 0.18 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_+_153197459 | 0.17 |
ENSRNOT00000023187
|
Rhd
|
Rh blood group, D antigen |
| chr1_-_164205222 | 0.16 |
ENSRNOT00000035774
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr5_+_138685624 | 0.16 |
ENSRNOT00000011867
|
Guca2a
|
guanylate cyclase activator 2A |
| chr8_+_50525091 | 0.15 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr7_+_138707426 | 0.14 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chr1_+_279798187 | 0.13 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr3_+_59153280 | 0.13 |
ENSRNOT00000002066
|
Cdca7
|
cell division cycle associated 7 |
| chr2_+_190073815 | 0.13 |
ENSRNOT00000015473
|
S100a8
|
S100 calcium binding protein A8 |
| chr16_-_61791091 | 0.12 |
ENSRNOT00000042003
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr19_-_10450186 | 0.12 |
ENSRNOT00000088048
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_-_35749464 | 0.11 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr2_+_116970344 | 0.09 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr1_+_199720038 | 0.09 |
ENSRNOT00000027297
|
Ahsp
|
alpha hemoglobin stabilizing protein |
| chr14_+_37130743 | 0.09 |
ENSRNOT00000031756
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr11_-_64952687 | 0.09 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chrX_-_78496847 | 0.09 |
ENSRNOT00000003265
|
Itm2a
|
integral membrane protein 2A |
| chr9_+_82647071 | 0.09 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr20_-_22459025 | 0.08 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chrX_+_112270986 | 0.08 |
ENSRNOT00000091441
ENSRNOT00000082652 |
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr11_-_60679555 | 0.07 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr17_-_84247038 | 0.07 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr17_-_21677477 | 0.07 |
ENSRNOT00000035448
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr4_+_119225040 | 0.06 |
ENSRNOT00000012365
|
Bmp10
|
bone morphogenetic protein 10 |
| chr9_-_88357182 | 0.06 |
ENSRNOT00000041176
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr5_+_25168295 | 0.06 |
ENSRNOT00000020245
|
Fsbp
|
fibrinogen silencer binding protein |
| chr9_-_82146874 | 0.06 |
ENSRNOT00000024190
|
Fev
|
FEV, ETS transcription factor |
| chr13_-_80745347 | 0.06 |
ENSRNOT00000041908
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr16_+_6970342 | 0.06 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr3_+_55910177 | 0.06 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr4_-_65780799 | 0.06 |
ENSRNOT00000079883
|
Atp6v0a4
|
ATPase H+ transporting V0 subunit a4 |
| chr13_-_53870428 | 0.06 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr10_-_55965216 | 0.06 |
ENSRNOT00000057058
|
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr13_+_48513008 | 0.06 |
ENSRNOT00000009742
|
Slc26a9
|
solute carrier family 26 member 9 |
| chr3_-_112985318 | 0.05 |
ENSRNOT00000015556
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr7_+_71023976 | 0.05 |
ENSRNOT00000005679
|
Tac3
|
tachykinin 3 |
| chr6_+_7961413 | 0.05 |
ENSRNOT00000007638
ENSRNOT00000061871 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr9_-_23493081 | 0.05 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chrX_-_13601069 | 0.05 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr8_-_117820413 | 0.05 |
ENSRNOT00000075819
|
Tma7
|
translation machinery associated 7 homolog |
| chr7_-_143538579 | 0.05 |
ENSRNOT00000081518
|
Krt79
|
keratin 79 |
| chr2_-_123851854 | 0.05 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr10_-_102227554 | 0.05 |
ENSRNOT00000078035
|
AC096468.1
|
|
| chr17_+_9639330 | 0.04 |
ENSRNOT00000018232
|
Dok3
|
docking protein 3 |
| chr6_+_137164535 | 0.04 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr3_-_72447801 | 0.04 |
ENSRNOT00000088815
|
P2rx3
|
purinergic receptor P2X 3 |
| chr8_+_22047697 | 0.04 |
ENSRNOT00000067741
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr9_-_85617954 | 0.04 |
ENSRNOT00000077331
|
Serpine2
|
serpin family E member 2 |
| chr6_+_83083740 | 0.04 |
ENSRNOT00000007600
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr20_-_2191640 | 0.04 |
ENSRNOT00000001016
|
Trim10
|
tripartite motif-containing 10 |
| chr7_+_117967818 | 0.04 |
ENSRNOT00000073724
|
LOC100360117
|
ribosomal protein L8-like |
| chr4_-_170932618 | 0.04 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr14_-_18862407 | 0.04 |
ENSRNOT00000003823
|
Cxcl6
|
C-X-C motif chemokine ligand 6 |
| chr18_-_19275273 | 0.04 |
ENSRNOT00000041707
|
LOC102547344
|
lateral signaling target protein 2 homolog |
| chr7_-_58149061 | 0.04 |
ENSRNOT00000005157
|
Tph2
|
tryptophan hydroxylase 2 |
| chr1_+_142883040 | 0.04 |
ENSRNOT00000015898
|
Alpk3
|
alpha-kinase 3 |
| chr15_-_61772516 | 0.03 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr5_+_133864798 | 0.03 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr10_-_15610826 | 0.03 |
ENSRNOT00000027851
|
Hbz
|
hemoglobin subunit zeta |
| chr9_-_105545854 | 0.03 |
ENSRNOT00000056434
|
RGD1560925
|
similar to 2610034M16Rik protein |
| chr1_+_32199810 | 0.03 |
ENSRNOT00000036714
|
Slc6a19
|
solute carrier family 6 member 19 |
| chr1_-_89094530 | 0.03 |
ENSRNOT00000031873
|
Etv2
|
ets variant 2 |
| chr2_-_192027225 | 0.03 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr15_+_33069382 | 0.03 |
ENSRNOT00000060193
|
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr2_-_196479770 | 0.03 |
ENSRNOT00000028693
|
Anxa9
|
annexin A9 |
| chr8_-_48672732 | 0.03 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr10_-_90415070 | 0.03 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr15_+_59678165 | 0.03 |
ENSRNOT00000074868
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr12_+_22274828 | 0.03 |
ENSRNOT00000001914
|
Epo
|
erythropoietin |
| chr19_-_14945302 | 0.03 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr20_-_5806097 | 0.03 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr8_+_39164916 | 0.02 |
ENSRNOT00000011309
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr2_-_60657712 | 0.02 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr2_+_189423559 | 0.02 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr11_+_61605937 | 0.02 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
| chr10_+_108132105 | 0.02 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr10_+_70417108 | 0.02 |
ENSRNOT00000079325
|
Slfn4
|
schlafen 4 |
| chr1_+_42121636 | 0.02 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr7_+_127964752 | 0.02 |
ENSRNOT00000038168
|
RGD1560617
|
hypothetical gene supported by NM_053561; AF062594 |
| chr2_-_28799266 | 0.02 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr2_-_189602143 | 0.02 |
ENSRNOT00000032062
|
Creb3l4
|
cAMP responsive element binding protein 3-like 4 |
| chr9_+_82700468 | 0.02 |
ENSRNOT00000027227
|
Inha
|
inhibin alpha subunit |
| chr9_+_52023295 | 0.02 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr1_+_20331976 | 0.02 |
ENSRNOT00000070858
|
Tmem200a
|
transmembrane protein 200A |
| chr1_+_17602281 | 0.02 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr20_-_27657983 | 0.02 |
ENSRNOT00000057313
|
Fam26e
|
family with sequence similarity 26, member E |
| chr18_-_62476700 | 0.02 |
ENSRNOT00000048429
|
Cycs
|
cytochrome c, somatic |
| chr4_+_51661654 | 0.02 |
ENSRNOT00000087359
ENSRNOT00000072470 |
LOC100909784
|
leiomodin-2-like |
| chrX_+_53053609 | 0.02 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr15_-_28696122 | 0.02 |
ENSRNOT00000060467
|
Rab2b
|
RAB2B, member RAS oncogene family |
| chr2_-_198002625 | 0.02 |
ENSRNOT00000091888
|
BC028528
|
cDNA sequence BC028528 |
| chr6_-_127653124 | 0.02 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr11_-_782954 | 0.02 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chrX_+_31054394 | 0.02 |
ENSRNOT00000037439
|
LOC688526
|
similar to ribonucleic acid binding protein S1 |
| chr15_-_45524582 | 0.02 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr1_+_243375981 | 0.02 |
ENSRNOT00000080689
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr6_-_26385761 | 0.02 |
ENSRNOT00000073228
|
Gckr
|
glucokinase regulator |
| chr14_-_86146744 | 0.02 |
ENSRNOT00000019335
|
Myl7
|
myosin light chain 7 |
| chrX_+_20351486 | 0.02 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr5_-_28130803 | 0.02 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr10_+_70411738 | 0.01 |
ENSRNOT00000078046
|
Slfn4
|
schlafen 4 |
| chr2_+_198721724 | 0.01 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr5_+_33097654 | 0.01 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chrX_+_106390759 | 0.01 |
ENSRNOT00000049375
|
Bhlhb9
|
basic helix-loop-helix domain containing, class B, 9 |
| chr4_-_165541314 | 0.01 |
ENSRNOT00000013833
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr20_+_31265483 | 0.01 |
ENSRNOT00000079584
ENSRNOT00000000674 |
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr3_+_93909156 | 0.01 |
ENSRNOT00000090365
|
Lmo2
|
LIM domain only 2 |
| chr11_-_78327282 | 0.01 |
ENSRNOT00000002636
|
Tp63
|
tumor protein p63 |
| chrX_+_83678339 | 0.01 |
ENSRNOT00000005997
|
Apool
|
apolipoprotein O-like |
| chr11_+_80736576 | 0.01 |
ENSRNOT00000047678
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr5_+_154077944 | 0.01 |
ENSRNOT00000031512
|
Il22ra1
|
interleukin 22 receptor subunit alpha 1 |
| chr4_-_70974006 | 0.01 |
ENSRNOT00000020975
|
Trpv5
|
transient receptor potential cation channel, subfamily V, member 5 |
| chr11_-_78341349 | 0.01 |
ENSRNOT00000068116
|
Tp63
|
tumor protein p63 |
| chr1_+_256955652 | 0.01 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr1_-_226732736 | 0.01 |
ENSRNOT00000072343
|
LOC108348129
|
pepsin F-like |
| chr6_-_8956276 | 0.01 |
ENSRNOT00000079027
|
Six2
|
SIX homeobox 2 |
| chr19_+_27404712 | 0.01 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr9_-_49950093 | 0.01 |
ENSRNOT00000023014
|
Fhl2
|
four and a half LIM domains 2 |
| chr14_-_45165207 | 0.01 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chrM_+_14136 | 0.01 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr3_+_159936856 | 0.01 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr1_-_242373764 | 0.01 |
ENSRNOT00000076544
|
Fam122a
|
family with sequence similarity 122A |
| chr2_+_42726809 | 0.01 |
ENSRNOT00000036860
|
AABR07008097.1
|
|
| chr13_-_61591139 | 0.01 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr2_-_209973205 | 0.01 |
ENSRNOT00000024593
|
Cym
|
chymosin |
| chr6_-_7421456 | 0.01 |
ENSRNOT00000006725
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr17_+_76008807 | 0.01 |
ENSRNOT00000070895
|
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr6_+_92229686 | 0.01 |
ENSRNOT00000046085
|
Atl1
|
atlastin GTPase 1 |
| chr1_+_20332371 | 0.00 |
ENSRNOT00000037259
|
Tmem200a
|
transmembrane protein 200A |
| chr3_+_72447969 | 0.00 |
ENSRNOT00000012022
|
Ssrp1
|
structure specific recognition protein 1 |
| chr15_-_49467174 | 0.00 |
ENSRNOT00000019209
|
Adam7
|
ADAM metallopeptidase domain 7 |
| chr19_+_41631715 | 0.00 |
ENSRNOT00000022749
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr9_-_88356716 | 0.00 |
ENSRNOT00000077503
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr6_-_7961207 | 0.00 |
ENSRNOT00000007174
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr18_+_27047382 | 0.00 |
ENSRNOT00000027691
ENSRNOT00000090264 |
Apc
|
APC, WNT signaling pathway regulator |
| chr20_-_5076539 | 0.00 |
ENSRNOT00000076253
ENSRNOT00000037613 |
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr4_-_7342410 | 0.00 |
ENSRNOT00000013058
|
Nos3
|
nitric oxide synthase 3 |
| chr19_-_37916813 | 0.00 |
ENSRNOT00000026585
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr15_+_28696378 | 0.00 |
ENSRNOT00000017476
|
Tox4
|
TOX high mobility group box family member 4 |
| chr2_+_208373154 | 0.00 |
ENSRNOT00000082943
ENSRNOT00000050538 |
LOC100911796
|
adenosine receptor A3-like |
| chr5_-_28131133 | 0.00 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr5_-_76632462 | 0.00 |
ENSRNOT00000085109
|
Susd1
|
sushi domain containing 1 |
| chr3_+_103753238 | 0.00 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr2_+_206454208 | 0.00 |
ENSRNOT00000026863
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr5_-_79691258 | 0.00 |
ENSRNOT00000072920
|
Tnfsf8
|
tumor necrosis factor superfamily member 8 |
| chr1_-_222272285 | 0.00 |
ENSRNOT00000028737
|
Fermt3
|
fermitin family member 3 |
| chr10_-_87232723 | 0.00 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr7_-_122247139 | 0.00 |
ENSRNOT00000064515
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr3_-_2453415 | 0.00 |
ENSRNOT00000079773
|
Slc34a3
|
solute carrier family 34 member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata2_Gata1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000041 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.2 | GO:1901258 | visceral motor neuron differentiation(GO:0021524) sensory system development(GO:0048880) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.1 | 0.2 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.0 | 0.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |