Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
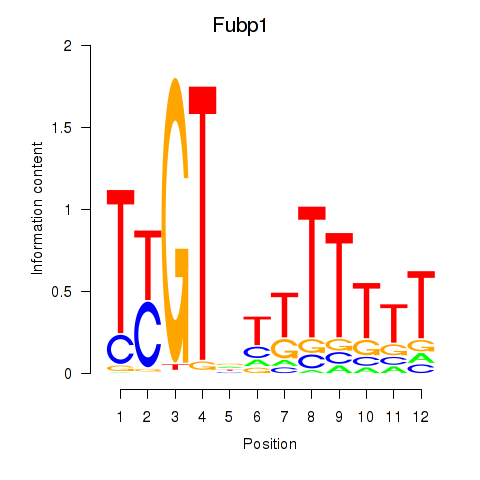
Results for Fubp1
Z-value: 0.32
Transcription factors associated with Fubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fubp1
|
ENSRNOG00000043370 | far upstream element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fubp1 | rn6_v1_chr2_+_257425679_257425679 | -0.41 | 4.9e-01 | Click! |
Activity profile of Fubp1 motif
Sorted Z-values of Fubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_46519431 | 0.15 |
ENSRNOT00000077765
|
AABR07045405.1
|
|
| chr15_-_30147793 | 0.14 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr20_+_6356423 | 0.12 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr6_-_146195819 | 0.11 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr16_+_72388880 | 0.11 |
ENSRNOT00000072459
|
LOC684871
|
similar to Protein C8orf4 (Thyroid cancer protein 1) (TC-1) |
| chr1_-_211923929 | 0.10 |
ENSRNOT00000054887
|
Nkx6-2
|
NK6 homeobox 2 |
| chr8_+_98755104 | 0.09 |
ENSRNOT00000056562
|
Zic4
|
Zic family member 4 |
| chr18_+_74006046 | 0.09 |
ENSRNOT00000086400
|
AABR07032622.1
|
|
| chr10_+_74724549 | 0.09 |
ENSRNOT00000009077
|
Tex14
|
testis expressed 14, intercellular bridge forming factor |
| chr10_-_97756521 | 0.08 |
ENSRNOT00000045902
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr7_+_120067379 | 0.06 |
ENSRNOT00000011270
|
Cdc42ep1
|
CDC42 effector protein 1 |
| chr1_-_24191908 | 0.06 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_-_114823500 | 0.06 |
ENSRNOT00000089870
|
NEWGENE_1562258
|
INO80 complex subunit B |
| chr15_+_17834635 | 0.06 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr8_-_55276070 | 0.06 |
ENSRNOT00000041555
|
LOC108351703
|
60S ribosomal protein L27a-like |
| chr4_-_51844331 | 0.05 |
ENSRNOT00000003593
|
Gpr37
|
G protein-coupled receptor 37 |
| chr18_+_51523758 | 0.05 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr18_-_59819113 | 0.05 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr3_-_11382004 | 0.04 |
ENSRNOT00000047921
ENSRNOT00000064039 |
Dnm1
|
dynamin 1 |
| chr1_+_225037737 | 0.04 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr1_+_264507985 | 0.04 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chr14_-_16276068 | 0.04 |
ENSRNOT00000002876
|
Ccng2
|
cyclin G2 |
| chr10_-_85825735 | 0.04 |
ENSRNOT00000055379
|
Fbxo47
|
F-box protein 47 |
| chr2_+_207262934 | 0.04 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr2_-_77784522 | 0.04 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr4_+_168689163 | 0.04 |
ENSRNOT00000049848
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr8_-_48692295 | 0.04 |
ENSRNOT00000065456
|
Vps11
|
VPS11 CORVET/HOPS core subunit |
| chr9_-_101107535 | 0.04 |
ENSRNOT00000080892
ENSRNOT00000071424 |
LOC100911516
|
coiled-coil-helix-coiled-coil-helix domain-containing protein 2, mitochondrial-like |
| chr1_-_91588609 | 0.04 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr9_+_46840992 | 0.03 |
ENSRNOT00000019415
|
Il1r2
|
interleukin 1 receptor type 2 |
| chr9_-_16406338 | 0.03 |
ENSRNOT00000073079
|
Tbcc
|
tubulin folding cofactor C |
| chr4_-_6978073 | 0.03 |
ENSRNOT00000045311
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr20_+_1749716 | 0.03 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr4_-_150185044 | 0.03 |
ENSRNOT00000019778
|
Csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr14_-_87701884 | 0.03 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr7_+_76980040 | 0.03 |
ENSRNOT00000050726
|
LOC108350501
|
40S ribosomal protein S29 |
| chr8_+_87211819 | 0.03 |
ENSRNOT00000086093
|
LOC100363289
|
LRRGT00022-like |
| chr19_-_53754602 | 0.03 |
ENSRNOT00000035651
|
Fam92b
|
family with sequence similarity 92, member B |
| chrX_-_29648359 | 0.03 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr20_+_27212724 | 0.03 |
ENSRNOT00000032600
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr13_-_94289333 | 0.03 |
ENSRNOT00000078195
|
Pld5
|
phospholipase D family, member 5 |
| chr5_-_100977902 | 0.02 |
ENSRNOT00000077429
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr1_+_264741911 | 0.02 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr10_+_15156207 | 0.02 |
ENSRNOT00000020363
|
Ccdc78
|
coiled-coil domain containing 78 |
| chr7_-_2623781 | 0.02 |
ENSRNOT00000004173
|
Spryd4
|
SPRY domain containing 4 |
| chr1_+_198067066 | 0.02 |
ENSRNOT00000084824
|
Sgf29
|
SAGA complex associated factor 29 |
| chr1_-_264756546 | 0.02 |
ENSRNOT00000020020
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr4_+_89536647 | 0.02 |
ENSRNOT00000058581
|
Tigd2
|
tigger transposable element derived 2 |
| chr7_-_60742660 | 0.02 |
ENSRNOT00000086116
|
Mdm2
|
MDM2 proto-oncogene |
| chr1_+_68436917 | 0.02 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chrX_+_105575759 | 0.02 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr4_+_168599331 | 0.01 |
ENSRNOT00000086719
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr1_-_219395623 | 0.01 |
ENSRNOT00000040261
|
Rpl35a
|
ribosomal protein L35a |
| chr12_-_7186873 | 0.01 |
ENSRNOT00000050438
|
LOC103690996
|
uncharacterized LOC103690996 |
| chr5_+_162808646 | 0.01 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr18_-_81807627 | 0.01 |
ENSRNOT00000020376
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr12_-_43576754 | 0.01 |
ENSRNOT00000042331
ENSRNOT00000081780 |
Med13l
|
mediator complex subunit 13-like |
| chr2_+_39314714 | 0.01 |
ENSRNOT00000083786
ENSRNOT00000047282 |
Smim15
|
small integral membrane protein 15 |
| chr17_-_63879166 | 0.01 |
ENSRNOT00000049391
ENSRNOT00000045183 |
Zmynd11
|
zinc finger, MYND-type containing 11 |
| chr1_-_216828581 | 0.01 |
ENSRNOT00000066943
ENSRNOT00000088856 |
Tnfrsf26
|
tumor necrosis factor receptor superfamily, member 26 |
| chr1_+_193076430 | 0.01 |
ENSRNOT00000086492
ENSRNOT00000017885 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr10_+_102167771 | 0.01 |
ENSRNOT00000003804
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr2_-_211831551 | 0.01 |
ENSRNOT00000039225
ENSRNOT00000090561 |
Fam102b
|
family with sequence similarity 102, member B |
| chr18_+_35850132 | 0.01 |
ENSRNOT00000067337
ENSRNOT00000075841 |
LOC103690064
|
m7GpppN-mRNA hydrolase-like |
| chr5_+_164923302 | 0.01 |
ENSRNOT00000042285
|
LOC108349682
|
60S ribosomal protein L35a |
| chr3_-_52447622 | 0.01 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr4_-_71870273 | 0.01 |
ENSRNOT00000077384
|
Olr801
|
olfactory receptor 801 |
| chr4_-_152883210 | 0.01 |
ENSRNOT00000056200
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr12_+_13715843 | 0.01 |
ENSRNOT00000042459
|
Actb
|
actin, beta |
| chr14_-_28536260 | 0.01 |
ENSRNOT00000059942
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr4_+_163293724 | 0.01 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr2_+_45104305 | 0.01 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr2_-_189333322 | 0.01 |
ENSRNOT00000073599
ENSRNOT00000071253 ENSRNOT00000076325 |
Hax1
|
HCLS1 associated protein X-1 |
| chr1_-_167210919 | 0.00 |
ENSRNOT00000027507
|
Chrna10
|
cholinergic receptor nicotinic alpha 10 subunit |
| chr14_+_89314176 | 0.00 |
ENSRNOT00000006765
|
Upp1
|
uridine phosphorylase 1 |
| chr4_-_120041238 | 0.00 |
ENSRNOT00000073799
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr2_+_114386019 | 0.00 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr12_-_1195591 | 0.00 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr13_-_26769374 | 0.00 |
ENSRNOT00000003768
|
Bcl2
|
BCL2, apoptosis regulator |
| chr15_+_47373120 | 0.00 |
ENSRNOT00000070815
|
Rp1l1
|
RP1 like 1 |
| chr6_+_126353223 | 0.00 |
ENSRNOT00000010385
|
Golga5
|
golgin A5 |
| chr3_-_161272460 | 0.00 |
ENSRNOT00000020740
|
Acot8
|
acyl-CoA thioesterase 8 |
| chr3_-_8955538 | 0.00 |
ENSRNOT00000040570
|
LOC686066
|
similar to 60S ribosomal protein L38 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.0 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.0 | 0.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |