Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Foxp1_Foxj2
Z-value: 0.50
Transcription factors associated with Foxp1_Foxj2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
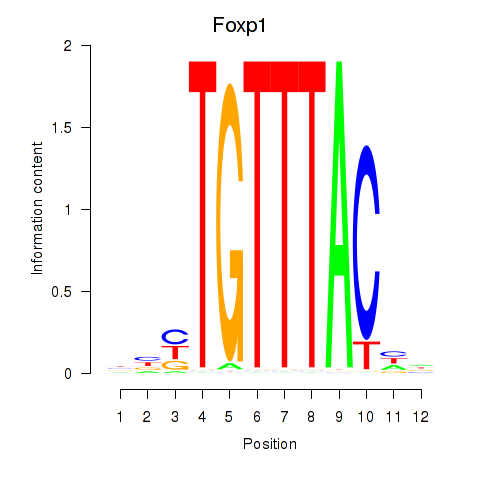
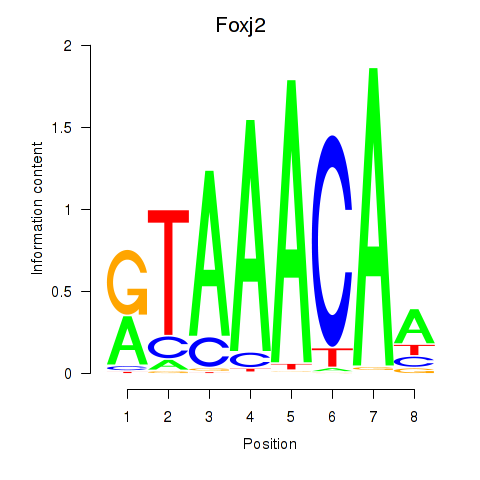
Foxp1
|
ENSRNOG00000009184 | forkhead box P1 |
|
Foxj2
|
ENSRNOG00000009000 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxp1 | rn6_v1_chr4_-_131694755_131694755 | 0.67 | 2.2e-01 | Click! |
| Foxj2 | rn6_v1_chr4_+_155654911_155654911 | 0.36 | 5.5e-01 | Click! |
Activity profile of Foxp1_Foxj2 motif
Sorted Z-values of Foxp1_Foxj2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_91054974 | 0.34 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr6_-_95502775 | 0.27 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr2_-_208225888 | 0.27 |
ENSRNOT00000054860
|
AABR07012775.1
|
|
| chr11_+_69739384 | 0.24 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_-_67520356 | 0.24 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr3_+_116072294 | 0.23 |
ENSRNOT00000071786
|
AABR07053613.1
|
|
| chrX_-_106607352 | 0.23 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr15_+_41937880 | 0.23 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr2_+_31378743 | 0.20 |
ENSRNOT00000050384
|
AABR07007853.1
|
|
| chr12_-_10391270 | 0.19 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr13_-_113817995 | 0.19 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chr10_-_103687425 | 0.18 |
ENSRNOT00000039284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr3_+_156886454 | 0.18 |
ENSRNOT00000022403
|
Lpin3
|
lipin 3 |
| chr19_-_21970103 | 0.18 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr7_+_23403891 | 0.17 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr4_+_41364441 | 0.17 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr3_+_147073160 | 0.16 |
ENSRNOT00000012690
|
Sdcbp2
|
syndecan binding protein 2 |
| chr9_+_94702129 | 0.16 |
ENSRNOT00000080930
|
Neu2
|
neuraminidase 2 |
| chr8_+_128087345 | 0.16 |
ENSRNOT00000019777
|
Acvr2b
|
activin A receptor type 2B |
| chr12_-_39396042 | 0.15 |
ENSRNOT00000001746
|
P2rx7
|
purinergic receptor P2X 7 |
| chr6_-_115513354 | 0.15 |
ENSRNOT00000005881
|
Ston2
|
stonin 2 |
| chr8_-_50245838 | 0.15 |
ENSRNOT00000035310
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr10_+_70262361 | 0.14 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_-_102919567 | 0.14 |
ENSRNOT00000072162
|
LOC100361143
|
ribosomal protein L30-like |
| chr4_+_115416580 | 0.14 |
ENSRNOT00000018303
|
Atp6v1b1
|
ATPase H+ transporting V1 subunit B1 |
| chr19_+_38601786 | 0.14 |
ENSRNOT00000087655
|
Zfp90
|
zinc finger protein 90 |
| chr13_-_1946508 | 0.13 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr10_+_86795787 | 0.13 |
ENSRNOT00000029021
|
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr13_+_50103189 | 0.13 |
ENSRNOT00000004078
|
Atp2b4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr4_+_88695590 | 0.13 |
ENSRNOT00000087835
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr14_+_104250617 | 0.12 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr1_-_254735548 | 0.12 |
ENSRNOT00000025258
|
Ankrd1
|
ankyrin repeat domain 1 |
| chr17_-_86657473 | 0.12 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr6_+_108167716 | 0.12 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr1_+_252409268 | 0.12 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr11_-_11585078 | 0.12 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr3_+_103773459 | 0.12 |
ENSRNOT00000079727
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr3_-_109044420 | 0.12 |
ENSRNOT00000007687
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr3_+_155297566 | 0.11 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr13_+_89597138 | 0.11 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr10_-_36419926 | 0.11 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr1_+_141488272 | 0.11 |
ENSRNOT00000034042
|
Wdr93
|
WD repeat domain 93 |
| chr14_+_71542057 | 0.11 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr2_+_41467064 | 0.11 |
ENSRNOT00000073231
|
AABR07008066.2
|
|
| chr10_-_97582188 | 0.11 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr17_+_86408151 | 0.11 |
ENSRNOT00000022734
|
Otud1
|
OTU deubiquitinase 1 |
| chr1_-_246110218 | 0.11 |
ENSRNOT00000077544
|
Rfx3
|
regulatory factor X3 |
| chr5_-_50075454 | 0.10 |
ENSRNOT00000011085
|
Orc3
|
origin recognition complex, subunit 3 |
| chr2_-_236823145 | 0.10 |
ENSRNOT00000073001
|
AABR07013425.1
|
|
| chr1_-_43638161 | 0.10 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr20_-_31598118 | 0.10 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr15_+_36865548 | 0.10 |
ENSRNOT00000076460
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr16_-_69280109 | 0.10 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr1_-_267203986 | 0.10 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr19_+_38162515 | 0.10 |
ENSRNOT00000027117
|
Slc7a6
|
solute carrier family 7 member 6 |
| chr17_-_21677477 | 0.10 |
ENSRNOT00000035448
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr13_-_84217332 | 0.10 |
ENSRNOT00000047488
ENSRNOT00000087096 |
Pou2f1
|
POU class 2 homeobox 1 |
| chr10_-_90501819 | 0.10 |
ENSRNOT00000050474
|
Gpatch8
|
G patch domain containing 8 |
| chr2_-_2779109 | 0.10 |
ENSRNOT00000017098
|
Rfesd
|
Rieske (Fe-S) domain containing |
| chr10_-_74298599 | 0.09 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr1_+_190671696 | 0.09 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr16_-_32299542 | 0.09 |
ENSRNOT00000072321
|
AABR07025299.1
|
|
| chr6_-_76608864 | 0.09 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr15_-_100348760 | 0.09 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr4_+_157594436 | 0.09 |
ENSRNOT00000029053
|
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr2_-_98610368 | 0.09 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr2_-_7651673 | 0.09 |
ENSRNOT00000085063
|
AC123427.1
|
|
| chr2_-_234296145 | 0.09 |
ENSRNOT00000014155
|
Elovl6
|
ELOVL fatty acid elongase 6 |
| chr4_+_168615890 | 0.09 |
ENSRNOT00000009324
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr12_-_51965779 | 0.09 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr1_-_188097530 | 0.09 |
ENSRNOT00000078825
|
Syt17
|
synaptotagmin 17 |
| chr3_-_90751055 | 0.09 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr6_-_75589257 | 0.09 |
ENSRNOT00000006030
|
Eapp
|
E2F-associated phosphoprotein |
| chrX_+_55331389 | 0.09 |
ENSRNOT00000075300
|
AABR07038567.1
|
|
| chr19_+_22450030 | 0.09 |
ENSRNOT00000021739
|
Neto2
|
neuropilin and tolloid like 2 |
| chr1_-_167911961 | 0.09 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr18_+_59748444 | 0.09 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr4_-_115516296 | 0.08 |
ENSRNOT00000019399
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr8_+_99880073 | 0.08 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr20_+_3823596 | 0.08 |
ENSRNOT00000087670
|
Rxrb
|
retinoid X receptor beta |
| chr3_+_101899068 | 0.08 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr2_-_185303610 | 0.08 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr1_-_170186462 | 0.08 |
ENSRNOT00000035798
|
Olr202
|
olfactory receptor 202 |
| chr7_+_121841855 | 0.08 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr14_-_46153212 | 0.08 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr8_+_104040934 | 0.08 |
ENSRNOT00000081204
|
Tfdp2
|
transcription factor Dp-2 |
| chr10_+_108132105 | 0.08 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr7_-_120882392 | 0.08 |
ENSRNOT00000056179
ENSRNOT00000056178 |
Fam227a
|
family with sequence similarity 227, member A |
| chr2_-_30846149 | 0.08 |
ENSRNOT00000025506
|
Slc30a5
|
solute carrier family 30 member 5 |
| chrX_+_115208863 | 0.08 |
ENSRNOT00000072094
|
AABR07040936.1
|
|
| chr1_+_255801694 | 0.08 |
ENSRNOT00000018878
|
AC103056.1
|
|
| chr5_+_118574801 | 0.08 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr6_+_29518416 | 0.08 |
ENSRNOT00000078660
|
AABR07063346.1
|
|
| chr7_-_130827152 | 0.08 |
ENSRNOT00000019406
|
Syt10
|
synaptotagmin 10 |
| chr8_-_104593625 | 0.08 |
ENSRNOT00000016625
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr3_+_113918629 | 0.08 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr7_+_71157664 | 0.08 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr10_-_110585376 | 0.08 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr1_-_235405831 | 0.08 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr7_-_140502441 | 0.08 |
ENSRNOT00000089544
|
Kmt2d
|
lysine methyltransferase 2D |
| chr5_-_127661028 | 0.07 |
ENSRNOT00000015466
|
Podn
|
podocan |
| chr10_-_27179254 | 0.07 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr6_-_108167185 | 0.07 |
ENSRNOT00000015545
|
Aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_+_45923222 | 0.07 |
ENSRNOT00000092976
ENSRNOT00000084454 ENSRNOT00000022939 |
Arid1b
|
AT-rich interaction domain 1B |
| chr9_-_20528879 | 0.07 |
ENSRNOT00000085293
|
AABR07066871.3
|
|
| chr4_-_40136061 | 0.07 |
ENSRNOT00000009752
|
Bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr6_-_76535517 | 0.07 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr2_+_3662763 | 0.07 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr18_+_27558089 | 0.07 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr7_-_118396728 | 0.07 |
ENSRNOT00000066431
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr1_-_163554839 | 0.07 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr15_-_108326907 | 0.07 |
ENSRNOT00000016848
|
Gpr18
|
G protein-coupled receptor 18 |
| chrX_+_15113878 | 0.07 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr3_+_120726906 | 0.07 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr7_+_143122269 | 0.07 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr9_+_111249351 | 0.07 |
ENSRNOT00000076729
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr8_+_91099491 | 0.07 |
ENSRNOT00000084852
|
Sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr17_+_81922329 | 0.07 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr3_+_64190973 | 0.07 |
ENSRNOT00000011342
|
AABR07052587.1
|
|
| chr19_+_33130303 | 0.07 |
ENSRNOT00000048673
|
LOC102550668
|
60S ribosomal protein L21-like |
| chr8_-_69466618 | 0.07 |
ENSRNOT00000042925
|
Ids
|
iduronate 2-sulfatase |
| chr16_-_25192675 | 0.07 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr1_-_31055453 | 0.07 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr20_-_4390436 | 0.07 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_+_42933077 | 0.07 |
ENSRNOT00000003361
|
Mfap3
|
microfibrillar-associated protein 3 |
| chr4_+_157438605 | 0.07 |
ENSRNOT00000079988
|
AC115420.4
|
|
| chr6_+_64649194 | 0.07 |
ENSRNOT00000039776
|
AABR07064102.1
|
|
| chr2_-_250923744 | 0.07 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr10_-_13826945 | 0.07 |
ENSRNOT00000012506
|
E4f1
|
E4F transcription factor 1 |
| chr6_+_21708487 | 0.07 |
ENSRNOT00000087358
|
AABR07063197.1
|
|
| chr12_-_11163981 | 0.07 |
ENSRNOT00000080154
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr18_-_40218225 | 0.07 |
ENSRNOT00000004723
|
Pggt1b
|
protein geranylgeranyltransferase type 1 subunit beta |
| chrX_-_105390580 | 0.07 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr2_-_232373409 | 0.07 |
ENSRNOT00000041479
|
AABR07013302.1
|
|
| chr6_+_135313008 | 0.07 |
ENSRNOT00000030864
|
Tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr2_+_205553163 | 0.07 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr7_-_98197087 | 0.06 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr15_+_1054937 | 0.06 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr8_-_107602263 | 0.06 |
ENSRNOT00000017658
|
Esyt3
|
extended synaptotagmin 3 |
| chr5_-_88629491 | 0.06 |
ENSRNOT00000058906
|
Tle1
|
transducin like enhancer of split 1 |
| chr5_-_7874909 | 0.06 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr10_+_73868943 | 0.06 |
ENSRNOT00000081012
|
Tubd1
|
tubulin, delta 1 |
| chr18_+_65155685 | 0.06 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr6_-_77421286 | 0.06 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr4_+_120671489 | 0.06 |
ENSRNOT00000029125
|
Mgll
|
monoglyceride lipase |
| chr2_+_118547190 | 0.06 |
ENSRNOT00000083676
ENSRNOT00000090301 ENSRNOT00000013410 |
Kcnmb2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr8_+_23398030 | 0.06 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr7_+_24638208 | 0.06 |
ENSRNOT00000009135
|
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr1_-_225631468 | 0.06 |
ENSRNOT00000072579
|
AABR07006232.1
|
|
| chr8_-_61519507 | 0.06 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr15_-_77736892 | 0.06 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr2_-_250232295 | 0.06 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr17_+_69617313 | 0.06 |
ENSRNOT00000073029
|
AABR07072562.1
|
|
| chr6_-_50786967 | 0.06 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr5_-_76756140 | 0.06 |
ENSRNOT00000022107
ENSRNOT00000089251 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr16_-_930527 | 0.06 |
ENSRNOT00000014787
|
Spin1
|
spindlin 1 |
| chr2_-_210943620 | 0.06 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr1_+_227757425 | 0.06 |
ENSRNOT00000032937
ENSRNOT00000049574 |
Ms4a6bl
|
membrane-spanning 4-domains, subfamily A, member 6B-like |
| chr18_+_63599425 | 0.06 |
ENSRNOT00000023145
|
Cep192
|
centrosomal protein 192 |
| chr1_+_15642153 | 0.06 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr1_-_1889236 | 0.06 |
ENSRNOT00000087944
|
AABR07000159.2
|
|
| chr8_-_53362006 | 0.06 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr10_-_74119009 | 0.06 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr6_+_86785771 | 0.06 |
ENSRNOT00000066702
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr3_+_46185360 | 0.06 |
ENSRNOT00000051344
|
LOC100361645
|
LRRGT00075-like |
| chr13_+_51384389 | 0.06 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr1_-_40972826 | 0.06 |
ENSRNOT00000060757
|
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr3_-_110517163 | 0.06 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr1_+_27476375 | 0.06 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr2_-_40386669 | 0.06 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr1_-_44615760 | 0.06 |
ENSRNOT00000022148
|
Nox3
|
NADPH oxidase 3 |
| chr6_-_109095557 | 0.06 |
ENSRNOT00000033493
|
Mlh3
|
mutL homolog 3 |
| chr10_-_74724472 | 0.06 |
ENSRNOT00000008846
|
Rad51c
|
RAD51 paralog C |
| chr4_-_11497531 | 0.06 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_+_104320743 | 0.06 |
ENSRNOT00000005684
|
Tmem94
|
transmembrane protein 94 |
| chr6_+_73358112 | 0.06 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr3_-_123607352 | 0.06 |
ENSRNOT00000051404
ENSRNOT00000028854 |
Adam33
|
ADAM metallopeptidase domain 33 |
| chr20_+_3558827 | 0.06 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_-_60795951 | 0.06 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr10_+_53570989 | 0.06 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr6_-_91581262 | 0.06 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr14_+_23611735 | 0.06 |
ENSRNOT00000031074
|
Cenpc
|
centromere protein C |
| chr9_-_32019205 | 0.06 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr10_+_68588789 | 0.06 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr9_+_61738471 | 0.06 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr15_-_13228607 | 0.06 |
ENSRNOT00000042010
ENSRNOT00000088214 |
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr2_+_234375315 | 0.06 |
ENSRNOT00000071270
|
LOC102549542
|
elongation of very long chain fatty acids protein 6-like |
| chr3_+_177351518 | 0.06 |
ENSRNOT00000023989
|
Pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr17_+_49417067 | 0.06 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr7_+_132857628 | 0.06 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr1_-_266830976 | 0.05 |
ENSRNOT00000027450
|
Pcgf6
|
polycomb group ring finger 6 |
| chr3_-_162035614 | 0.05 |
ENSRNOT00000047890
|
Zfp663
|
zinc finger protein 663 |
| chr5_-_160403373 | 0.05 |
ENSRNOT00000018393
|
Ctrc
|
chymotrypsin C |
| chr3_-_161299024 | 0.05 |
ENSRNOT00000021216
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr5_+_153260930 | 0.05 |
ENSRNOT00000023289
ENSRNOT00000082184 |
Rsrp1
|
arginine and serine rich protein 1 |
| chr1_+_15834779 | 0.05 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr2_+_54191538 | 0.05 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr7_-_68512397 | 0.05 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr12_+_31335372 | 0.05 |
ENSRNOT00000001242
ENSRNOT00000037296 |
Stx2
|
syntaxin 2 |
| chr1_-_98570949 | 0.05 |
ENSRNOT00000033648
|
Siglec5
|
sialic acid binding Ig-like lectin 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxp1_Foxj2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.2 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.2 | GO:1904170 | phospholipid transfer to membrane(GO:0006649) NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) |
| 0.0 | 0.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 0.0 | 0.1 | GO:0046340 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.0 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.0 | 0.1 | GO:2000468 | positive regulation of protein autoubiquitination(GO:1902499) regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.0 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:2000124 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.0 | 0.0 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0033364 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.0 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.0 | GO:1904404 | response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.0 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.0 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.0 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.0 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.0 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0004948 | calcitonin gene-related peptide receptor activity(GO:0001635) calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.0 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |