Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
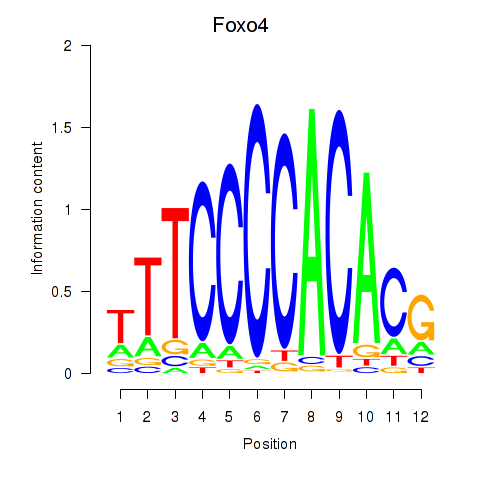
Results for Foxo4
Z-value: 0.22
Transcription factors associated with Foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo4
|
ENSRNOG00000033316 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo4 | rn6_v1_chrX_+_71155601_71155601 | -0.38 | 5.3e-01 | Click! |
Activity profile of Foxo4 motif
Sorted Z-values of Foxo4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_90049112 | 0.10 |
ENSRNOT00000028323
|
Pyy
|
peptide YY (mapped) |
| chr12_-_17186679 | 0.10 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr1_+_168957460 | 0.09 |
ENSRNOT00000090745
|
LOC103694857
|
hemoglobin subunit beta-2 |
| chr1_-_168972725 | 0.09 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr1_+_168945449 | 0.08 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr7_+_144052061 | 0.08 |
ENSRNOT00000020103
ENSRNOT00000091378 |
Amhr2
|
anti-Mullerian hormone receptor type 2 |
| chr5_-_139933764 | 0.06 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr3_+_2877293 | 0.05 |
ENSRNOT00000061855
|
Lcn5
|
lipocalin 5 |
| chr14_+_83510640 | 0.05 |
ENSRNOT00000089928
ENSRNOT00000082242 ENSRNOT00000025378 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr1_+_189665976 | 0.04 |
ENSRNOT00000033673
|
Lyrm1
|
LYR motif containing 1 |
| chr3_+_122544788 | 0.04 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr3_+_91870208 | 0.04 |
ENSRNOT00000040249
|
Rpl21
|
ribosomal protein L21 |
| chr4_-_70996395 | 0.03 |
ENSRNOT00000021485
|
Kel
|
Kell blood group, metallo-endopeptidase |
| chr20_+_26534535 | 0.03 |
ENSRNOT00000000423
|
Tmem167b
|
transmembrane protein 167B |
| chr1_+_226252226 | 0.03 |
ENSRNOT00000039369
|
AABR07006258.1
|
|
| chr1_+_215609645 | 0.03 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr7_-_66172360 | 0.03 |
ENSRNOT00000005537
|
Fam19a2
|
family with sequence similarity 19 member A2, C-C motif chemokine like |
| chr4_+_159622404 | 0.02 |
ENSRNOT00000078299
|
Fgf23
|
fibroblast growth factor 23 |
| chr15_-_28696122 | 0.02 |
ENSRNOT00000060467
|
Rab2b
|
RAB2B, member RAS oncogene family |
| chr9_+_67774150 | 0.02 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr1_-_80483487 | 0.02 |
ENSRNOT00000074418
|
Gemin7
|
gem (nuclear organelle) associated protein 7 |
| chr10_-_13081136 | 0.02 |
ENSRNOT00000005718
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr14_+_88549947 | 0.02 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr3_-_111037166 | 0.02 |
ENSRNOT00000017070
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr7_+_23913830 | 0.02 |
ENSRNOT00000006570
|
Rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr6_+_73345392 | 0.02 |
ENSRNOT00000088378
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr10_-_18090932 | 0.01 |
ENSRNOT00000064744
ENSRNOT00000006591 |
Npm1
|
nucleophosmin 1 |
| chr3_+_9643047 | 0.01 |
ENSRNOT00000035805
|
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr1_-_82236179 | 0.01 |
ENSRNOT00000027845
|
Cnfn
|
cornifelin |
| chr1_-_216828581 | 0.01 |
ENSRNOT00000066943
ENSRNOT00000088856 |
Tnfrsf26
|
tumor necrosis factor receptor superfamily, member 26 |
| chr19_-_869490 | 0.01 |
ENSRNOT00000080433
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr19_+_9436210 | 0.01 |
ENSRNOT00000074845
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr5_+_148577332 | 0.01 |
ENSRNOT00000016325
|
Snrnp40
|
small nuclear ribonucleoprotein U5 subunit 40 |
| chr4_+_71675383 | 0.01 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr12_+_24180937 | 0.01 |
ENSRNOT00000030497
|
Hip1
|
huntingtin interacting protein 1 |
| chr2_-_187426375 | 0.01 |
ENSRNOT00000025986
|
Naxe
|
NAD(P)HX epimerase |
| chr15_-_51834030 | 0.01 |
ENSRNOT00000024895
|
Ccar2
|
cell cycle and apoptosis regulator 2 |
| chr9_+_50966766 | 0.01 |
ENSRNOT00000076636
|
Ercc5
|
ERCC excision repair 5, endonuclease |
| chr19_+_46733633 | 0.01 |
ENSRNOT00000016307
|
Clec3a
|
C-type lectin domain family 3, member A |
| chr16_-_8207464 | 0.01 |
ENSRNOT00000026742
|
Dph3
|
diphthamide biosynthesis 3 |
| chr20_-_157861 | 0.01 |
ENSRNOT00000084461
|
RT1-CE10
|
RT1 class I, locus CE10 |
| chr4_-_66624912 | 0.01 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr14_+_83510278 | 0.01 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_+_11657370 | 0.01 |
ENSRNOT00000073840
|
Ak1
|
adenylate kinase 1 |
| chr8_+_94693290 | 0.01 |
ENSRNOT00000013908
|
Cyb5r4
|
cytochrome b5 reductase 4 |
| chr20_+_5414448 | 0.01 |
ENSRNOT00000078972
ENSRNOT00000080900 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr5_-_148577292 | 0.00 |
ENSRNOT00000017156
|
Zcchc17
|
zinc finger CCHC-type containing 17 |
| chr10_+_93811350 | 0.00 |
ENSRNOT00000077280
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr6_+_126623015 | 0.00 |
ENSRNOT00000010672
|
Tmem251
|
transmembrane protein 251 |
| chr10_+_93811505 | 0.00 |
ENSRNOT00000081937
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr10_+_63098116 | 0.00 |
ENSRNOT00000030749
|
Nsrp1
|
nuclear speckle splicing regulatory protein 1 |
| chr3_+_149144795 | 0.00 |
ENSRNOT00000015482
|
Dnmt3b
|
DNA methyltransferase 3 beta |
| chr9_+_42620006 | 0.00 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |