Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
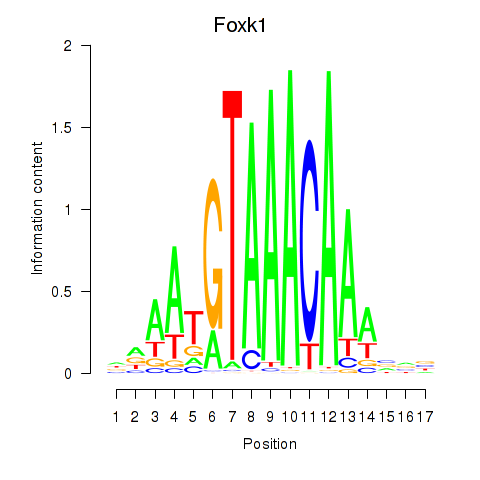
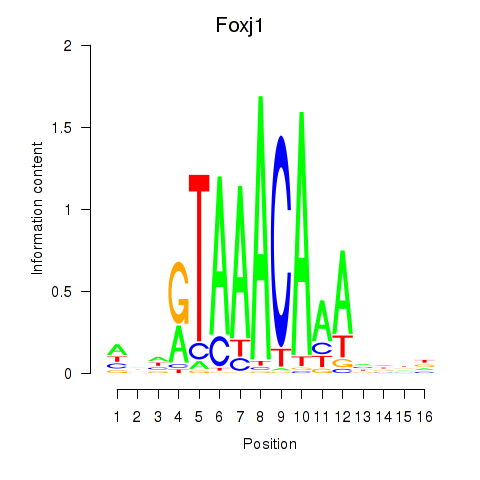
Results for Foxk1_Foxj1
Z-value: 0.62
Transcription factors associated with Foxk1_Foxj1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxk1
|
ENSRNOG00000001104 | forkhead box K1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxk1 | rn6_v1_chr12_-_14244316_14244316 | -0.09 | 8.8e-01 | Click! |
Activity profile of Foxk1_Foxj1 motif
Sorted Z-values of Foxk1_Foxj1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_12346117 | 0.28 |
ENSRNOT00000074599
|
AABR07066677.1
|
|
| chr1_+_141488272 | 0.25 |
ENSRNOT00000034042
|
Wdr93
|
WD repeat domain 93 |
| chr2_+_31378743 | 0.23 |
ENSRNOT00000050384
|
AABR07007853.1
|
|
| chr15_+_36865548 | 0.22 |
ENSRNOT00000076460
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr3_-_10153554 | 0.21 |
ENSRNOT00000093246
|
Exosc2
|
exosome component 2 |
| chr8_-_43480777 | 0.19 |
ENSRNOT00000060074
|
LOC688473
|
similar to 40S ribosomal protein S2 |
| chr8_-_43534620 | 0.17 |
ENSRNOT00000035386
|
LOC688473
|
similar to 40S ribosomal protein S2 |
| chr12_-_10391270 | 0.16 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr13_+_91054974 | 0.16 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr2_-_34963207 | 0.15 |
ENSRNOT00000017870
|
Fam159b
|
family with sequence similarity 159, member B |
| chr1_+_29432152 | 0.15 |
ENSRNOT00000019436
ENSRNOT00000083276 |
Trmt11
|
tRNA methyltransferase 11 homolog |
| chr18_-_410098 | 0.14 |
ENSRNOT00000084138
|
LOC102546764
|
cx9C motif-containing protein 4-like |
| chr4_-_176528110 | 0.14 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr6_-_95502775 | 0.13 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr17_-_15484269 | 0.13 |
ENSRNOT00000020648
|
Omd
|
osteomodulin |
| chr5_-_50075454 | 0.12 |
ENSRNOT00000011085
|
Orc3
|
origin recognition complex, subunit 3 |
| chr10_+_53570989 | 0.12 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr10_-_44746549 | 0.12 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr15_+_41937880 | 0.12 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr10_+_42933077 | 0.12 |
ENSRNOT00000003361
|
Mfap3
|
microfibrillar-associated protein 3 |
| chr5_-_62187930 | 0.11 |
ENSRNOT00000011787
|
Coro2a
|
coronin 2A |
| chr1_+_228684136 | 0.11 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr13_+_80125391 | 0.11 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr2_-_3124543 | 0.11 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr17_-_21677477 | 0.11 |
ENSRNOT00000035448
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr19_+_33130303 | 0.11 |
ENSRNOT00000048673
|
LOC102550668
|
60S ribosomal protein L21-like |
| chr15_-_44442875 | 0.11 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr14_-_3351553 | 0.10 |
ENSRNOT00000061556
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr2_-_232373409 | 0.10 |
ENSRNOT00000041479
|
AABR07013302.1
|
|
| chr6_+_27887797 | 0.10 |
ENSRNOT00000015827
|
Asxl2
|
additional sex combs like 2, transcriptional regulator |
| chrX_+_111122552 | 0.10 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chrX_+_10430847 | 0.09 |
ENSRNOT00000047936
|
Rpl21
|
ribosomal protein L21 |
| chr2_-_165591110 | 0.09 |
ENSRNOT00000091140
|
Ift80
|
intraflagellar transport 80 |
| chr1_-_84732242 | 0.09 |
ENSRNOT00000025586
|
LOC102553760
|
zinc finger protein 59-like |
| chr10_+_68588789 | 0.09 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr3_+_147073160 | 0.09 |
ENSRNOT00000012690
|
Sdcbp2
|
syndecan binding protein 2 |
| chr13_-_97838228 | 0.09 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr3_+_91870208 | 0.09 |
ENSRNOT00000040249
|
Rpl21
|
ribosomal protein L21 |
| chr16_-_32299542 | 0.09 |
ENSRNOT00000072321
|
AABR07025299.1
|
|
| chr20_-_4863011 | 0.09 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr4_+_123801174 | 0.08 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr13_-_98480419 | 0.08 |
ENSRNOT00000086306
|
Coq8a
|
coenzyme Q8A |
| chr7_+_141642777 | 0.08 |
ENSRNOT00000079811
|
AABR07058884.2
|
|
| chr8_+_32604365 | 0.08 |
ENSRNOT00000071210
|
AABR07069587.1
|
|
| chr9_-_65693822 | 0.08 |
ENSRNOT00000038431
|
Als2cr12
|
amyotrophic lateral sclerosis 2 chromosome region, candidate 12 |
| chr2_+_113007549 | 0.08 |
ENSRNOT00000017758
|
Tnfsf10
|
tumor necrosis factor superfamily member 10 |
| chr3_+_155269347 | 0.08 |
ENSRNOT00000021308
|
Fam83d
|
family with sequence similarity 83, member D |
| chr5_-_83648044 | 0.08 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr3_+_113918629 | 0.08 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr2_-_34419428 | 0.08 |
ENSRNOT00000048123
|
AABR07007905.1
|
|
| chr13_+_89597138 | 0.08 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr1_+_227592635 | 0.08 |
ENSRNOT00000072135
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr2_-_40386669 | 0.08 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr3_-_138708332 | 0.08 |
ENSRNOT00000010678
|
Rbbp9
|
RB binding protein 9, serine hydrolase |
| chr5_-_104980641 | 0.07 |
ENSRNOT00000071192
|
Haus6
|
HAUS augmin-like complex, subunit 6 |
| chr12_-_41237209 | 0.07 |
ENSRNOT00000088023
|
Oas1b
|
2-5 oligoadenylate synthetase 1B |
| chr5_+_50075527 | 0.07 |
ENSRNOT00000011688
|
Rars2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr9_+_12420368 | 0.07 |
ENSRNOT00000071620
|
AABR07066693.1
|
|
| chr2_-_208225888 | 0.07 |
ENSRNOT00000054860
|
AABR07012775.1
|
|
| chr1_-_40972826 | 0.07 |
ENSRNOT00000060757
|
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr11_-_15858281 | 0.07 |
ENSRNOT00000090586
|
AABR07033287.1
|
|
| chr7_+_138707426 | 0.07 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chr3_-_46726946 | 0.07 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr12_+_19680712 | 0.07 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr10_+_108132105 | 0.07 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr2_-_27287605 | 0.07 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr20_+_8484311 | 0.07 |
ENSRNOT00000050041
|
LOC100364116
|
ribosomal protein S20-like |
| chr19_-_21970103 | 0.07 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr7_-_10588202 | 0.07 |
ENSRNOT00000086920
|
AABR07055826.1
|
|
| chr9_+_77320726 | 0.07 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr9_-_7891514 | 0.07 |
ENSRNOT00000072684
|
Pot1b
|
protection of telomeres 1B |
| chr7_+_29909120 | 0.07 |
ENSRNOT00000049362
|
AABR07056556.1
|
|
| chr14_-_8600512 | 0.07 |
ENSRNOT00000092537
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr16_-_69280109 | 0.07 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr20_+_40778927 | 0.07 |
ENSRNOT00000001081
|
Smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr5_-_24560063 | 0.07 |
ENSRNOT00000037869
|
Dpy19l4
|
dpy-19-like 4 (C. elegans) |
| chrX_+_65040775 | 0.07 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr5_+_128190708 | 0.07 |
ENSRNOT00000012492
|
Orc1
|
origin recognition complex, subunit 1 |
| chr15_+_30557388 | 0.07 |
ENSRNOT00000072978
|
AABR07017745.4
|
|
| chr8_+_23398030 | 0.07 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr10_-_36419926 | 0.07 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr9_+_111249351 | 0.06 |
ENSRNOT00000076729
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr6_-_47848026 | 0.06 |
ENSRNOT00000011048
ENSRNOT00000090017 |
Allc
|
allantoicase |
| chr17_-_1648973 | 0.06 |
ENSRNOT00000036212
|
Slc35d2
|
solute carrier family 35 member D2 |
| chr10_+_3655038 | 0.06 |
ENSRNOT00000045047
|
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr20_-_4863198 | 0.06 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr3_+_60024013 | 0.06 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr8_-_127900463 | 0.06 |
ENSRNOT00000078303
|
Slc22a13
|
solute carrier family 22 member 13 |
| chr16_+_22361998 | 0.06 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr2_-_53827175 | 0.06 |
ENSRNOT00000078158
|
RGD1305938
|
similar to expressed sequence AW549877 |
| chr4_+_25635765 | 0.06 |
ENSRNOT00000009340
|
Gtpbp10
|
GTP binding protein 10 |
| chr3_-_471634 | 0.06 |
ENSRNOT00000081816
|
Spopl
|
speckle type BTB/POZ protein like |
| chr19_+_43338166 | 0.06 |
ENSRNOT00000091720
ENSRNOT00000084739 |
Fuk
|
fucokinase |
| chr6_-_107080524 | 0.06 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr18_-_81682206 | 0.06 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr10_-_95262024 | 0.06 |
ENSRNOT00000066487
|
Bptf
|
bromodomain PHD finger transcription factor |
| chr18_+_86116044 | 0.06 |
ENSRNOT00000058160
ENSRNOT00000076159 |
Rttn
|
rotatin |
| chr4_-_155923079 | 0.06 |
ENSRNOT00000013308
|
Clec4a3
|
C-type lectin domain family 4, member A3 |
| chr6_-_102472926 | 0.06 |
ENSRNOT00000079351
|
Zfyve26
|
zinc finger FYVE-type containing 26 |
| chr7_+_91384187 | 0.06 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr5_-_160403373 | 0.06 |
ENSRNOT00000018393
|
Ctrc
|
chymotrypsin C |
| chr17_-_21739408 | 0.06 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr20_+_46707362 | 0.06 |
ENSRNOT00000071045
|
AABR07045411.1
|
|
| chr2_+_195940715 | 0.06 |
ENSRNOT00000042985
|
LOC100363268
|
rCG31129-like |
| chr20_+_2088533 | 0.06 |
ENSRNOT00000001012
ENSRNOT00000079021 |
Znrd1
|
zinc ribbon domain containing 1 |
| chr4_+_178617401 | 0.06 |
ENSRNOT00000042619
|
AABR07062466.1
|
|
| chr9_-_95143092 | 0.06 |
ENSRNOT00000064171
ENSRNOT00000085325 |
Usp40
|
ubiquitin specific peptidase 40 |
| chr7_-_123586919 | 0.06 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr6_+_95826546 | 0.06 |
ENSRNOT00000043587
|
AABR07064810.1
|
|
| chr3_-_24601063 | 0.06 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr17_-_15478343 | 0.06 |
ENSRNOT00000078029
|
Omd
|
osteomodulin |
| chr4_-_67520356 | 0.05 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr18_+_30840868 | 0.05 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr6_-_93562314 | 0.05 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr8_-_48597867 | 0.05 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chr3_-_80012750 | 0.05 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_-_66237501 | 0.05 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chrX_-_15627235 | 0.05 |
ENSRNOT00000013369
|
Wdr45
|
WD repeat domain 45 |
| chr4_+_139670092 | 0.05 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr8_-_22937909 | 0.05 |
ENSRNOT00000015684
|
Tmem205
|
transmembrane protein 205 |
| chr3_-_71798531 | 0.05 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr8_+_99880073 | 0.05 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr3_-_101474890 | 0.05 |
ENSRNOT00000091869
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr5_+_61474000 | 0.05 |
ENSRNOT00000013930
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr17_-_70481750 | 0.05 |
ENSRNOT00000025252
|
Il15ra
|
interleukin 15 receptor subunit alpha |
| chr4_-_98995590 | 0.05 |
ENSRNOT00000008698
|
Thnsl2
|
threonine synthase-like 2 |
| chr4_+_158224000 | 0.05 |
ENSRNOT00000084240
ENSRNOT00000078495 |
Ano2
|
anoctamin 2 |
| chr4_-_120414118 | 0.05 |
ENSRNOT00000072795
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr15_-_25505691 | 0.05 |
ENSRNOT00000074488
|
LOC100911492
|
homeobox protein OTX2-like |
| chr7_-_68512397 | 0.05 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr9_+_73528681 | 0.05 |
ENSRNOT00000084340
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr1_-_246110218 | 0.05 |
ENSRNOT00000077544
|
Rfx3
|
regulatory factor X3 |
| chr5_-_127661028 | 0.05 |
ENSRNOT00000015466
|
Podn
|
podocan |
| chr6_-_51018050 | 0.05 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chrM_+_2740 | 0.05 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr20_-_3372070 | 0.05 |
ENSRNOT00000080770
|
Dhx16
|
DEAH-box helicase 16 |
| chr6_+_64649194 | 0.05 |
ENSRNOT00000039776
|
AABR07064102.1
|
|
| chr17_+_86066833 | 0.05 |
ENSRNOT00000022661
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr3_+_28416954 | 0.05 |
ENSRNOT00000043533
|
Kynu
|
kynureninase |
| chr14_-_46529375 | 0.05 |
ENSRNOT00000077306
ENSRNOT00000078682 |
LOC257642
|
rRNA promoter binding protein |
| chr9_-_55256340 | 0.05 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr2_+_189615948 | 0.05 |
ENSRNOT00000092144
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr12_-_11808977 | 0.05 |
ENSRNOT00000071795
|
AABR07035375.1
|
|
| chr3_-_119611136 | 0.05 |
ENSRNOT00000016157
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr11_-_27080701 | 0.05 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr19_+_55300395 | 0.05 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr18_+_40883224 | 0.05 |
ENSRNOT00000039733
|
Lvrn
|
laeverin |
| chr11_+_69739384 | 0.05 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr7_+_13151592 | 0.05 |
ENSRNOT00000010203
|
Vom2r53
|
vomeronasal 2 receptor, 53 |
| chr15_-_52443055 | 0.05 |
ENSRNOT00000087450
|
Xpo7
|
exportin 7 |
| chr17_-_8925503 | 0.05 |
ENSRNOT00000015163
|
Catsper3
|
cation channel, sperm associated 3 |
| chr20_+_25990656 | 0.05 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr8_-_61519507 | 0.04 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr1_+_61268248 | 0.04 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr13_-_84217332 | 0.04 |
ENSRNOT00000047488
ENSRNOT00000087096 |
Pou2f1
|
POU class 2 homeobox 1 |
| chr12_+_45319501 | 0.04 |
ENSRNOT00000090630
ENSRNOT00000041732 |
RGD1561114
|
similar to hypothetical protein 4930474N05 |
| chr9_+_42315682 | 0.04 |
ENSRNOT00000071995
|
AABR07067388.1
|
|
| chr4_+_77623538 | 0.04 |
ENSRNOT00000059263
|
Zfp956
|
zinc finger protein 956 |
| chr6_+_80108655 | 0.04 |
ENSRNOT00000006137
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr1_+_254974789 | 0.04 |
ENSRNOT00000025020
|
Pcgf5
|
polycomb group ring finger 5 |
| chr14_-_66978499 | 0.04 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr1_+_227217552 | 0.04 |
ENSRNOT00000074966
|
LOC100911403
|
membrane-spanning 4-domains subfamily A member 4A-like |
| chr13_-_61003744 | 0.04 |
ENSRNOT00000005163
|
Rgs13
|
regulator of G-protein signaling 13 |
| chr5_+_118574801 | 0.04 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr13_-_71276497 | 0.04 |
ENSRNOT00000029773
|
Rgsl1
|
regulator of G-protein signaling like 1 |
| chr12_-_20660304 | 0.04 |
ENSRNOT00000091323
|
RGD1561730
|
similar to cell surface receptor FDFACT |
| chr5_-_147635789 | 0.04 |
ENSRNOT00000037106
|
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr13_-_91776397 | 0.04 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr10_-_37130921 | 0.04 |
ENSRNOT00000006006
|
Trappc2b
|
trafficking protein particle complex 2B |
| chr5_-_50362344 | 0.04 |
ENSRNOT00000035808
|
Zfp292
|
zinc finger protein 292 |
| chr10_+_39875371 | 0.04 |
ENSRNOT00000013481
|
Rapgef6
|
Rap guanine nucleotide exchange factor 6 |
| chrX_-_123788898 | 0.04 |
ENSRNOT00000009123
|
Akap14
|
A-kinase anchoring protein 14 |
| chr15_+_2526368 | 0.04 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chr13_-_78521911 | 0.04 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr17_+_78911581 | 0.04 |
ENSRNOT00000085104
|
AC128960.2
|
|
| chr2_-_98610368 | 0.04 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr6_+_93462852 | 0.04 |
ENSRNOT00000089473
|
Arid4a
|
AT-rich interaction domain 4A |
| chr1_+_190852985 | 0.04 |
ENSRNOT00000040963
|
Polr3e
|
RNA polymerase III subunit E |
| chr9_-_49448168 | 0.04 |
ENSRNOT00000059478
|
AABR07067499.1
|
|
| chr6_-_51257625 | 0.04 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
| chr6_-_78549669 | 0.04 |
ENSRNOT00000009940
|
Foxa1
|
forkhead box A1 |
| chr8_-_26345754 | 0.04 |
ENSRNOT00000091773
|
AC094212.1
|
triosephosphate isomerase |
| chr20_-_38985036 | 0.04 |
ENSRNOT00000001066
|
Serinc1
|
serine incorporator 1 |
| chr8_+_99632803 | 0.04 |
ENSRNOT00000087190
|
Plscr1
|
phospholipid scramblase 1 |
| chr3_+_46185360 | 0.04 |
ENSRNOT00000051344
|
LOC100361645
|
LRRGT00075-like |
| chr16_-_25192675 | 0.04 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr7_+_11490852 | 0.04 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr17_+_47241017 | 0.04 |
ENSRNOT00000092193
|
Gpr141
|
G protein-coupled receptor 141 |
| chr10_-_27179900 | 0.04 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr1_-_67094567 | 0.04 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr18_-_61788859 | 0.04 |
ENSRNOT00000034075
ENSRNOT00000034069 |
Ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr15_-_89339323 | 0.04 |
ENSRNOT00000013297
|
Rbm26
|
RNA binding motif protein 26 |
| chrX_+_123751293 | 0.04 |
ENSRNOT00000089883
|
Nkap
|
NFKB activating protein |
| chr5_-_160383782 | 0.04 |
ENSRNOT00000018349
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr9_-_91100482 | 0.04 |
ENSRNOT00000064642
|
LOC100911860
|
ubiquitin carboxyl-terminal hydrolase 40-like |
| chr3_-_120157866 | 0.04 |
ENSRNOT00000020367
|
LOC100909998
|
zinc finger protein 2-like |
| chr15_-_37056892 | 0.04 |
ENSRNOT00000047731
|
Zmym5
|
zinc finger MYM-type containing 5 |
| chr3_-_61488696 | 0.04 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr14_-_46657975 | 0.04 |
ENSRNOT00000087247
|
LOC257642
|
rRNA promoter binding protein |
| chr5_+_60850852 | 0.04 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxk1_Foxj1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071049 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.0 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0051790 | threonine metabolic process(GO:0006566) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0004948 | calcitonin gene-related peptide receptor activity(GO:0001635) calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.0 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.0 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |