Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Foxg1
Z-value: 0.27
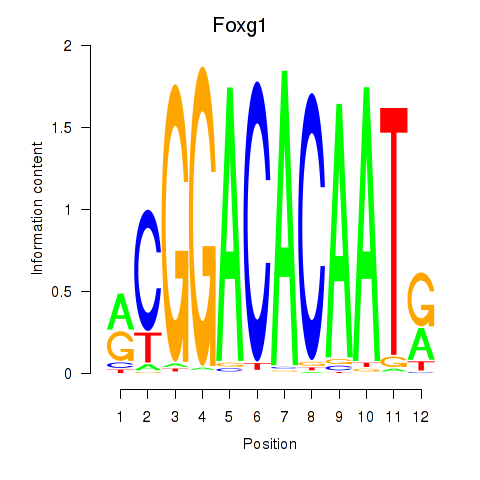
Transcription factors associated with Foxg1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxg1
|
ENSRNOG00000047891 | forkhead box G1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxg1 | rn6_v1_chr6_+_69971227_69971227 | 0.89 | 4.2e-02 | Click! |
Activity profile of Foxg1 motif
Sorted Z-values of Foxg1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_114866768 | 0.19 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr5_+_172364421 | 0.13 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr7_-_126465723 | 0.11 |
ENSRNOT00000021196
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr2_-_231521052 | 0.10 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr1_-_92119951 | 0.10 |
ENSRNOT00000018153
ENSRNOT00000092121 |
Zfp507
|
zinc finger protein 507 |
| chr8_+_114867062 | 0.09 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr14_-_112946204 | 0.07 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr6_-_29975730 | 0.07 |
ENSRNOT00000075574
|
Wdcp
|
WD repeat and coiled coil containing |
| chr1_-_164659992 | 0.06 |
ENSRNOT00000024281
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr3_-_148312791 | 0.06 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr5_-_20189721 | 0.05 |
ENSRNOT00000014661
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr13_-_95348913 | 0.05 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr9_-_80167033 | 0.04 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr10_+_13084304 | 0.04 |
ENSRNOT00000066310
|
RGD1561157
|
RGD1561157 |
| chr9_+_6970507 | 0.04 |
ENSRNOT00000079488
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr14_-_106864892 | 0.04 |
ENSRNOT00000090664
|
Otx1
|
orthodenticle homeobox 1 |
| chr3_-_22500765 | 0.04 |
ENSRNOT00000025443
|
Dennd1a
|
DENN domain containing 1A |
| chr15_-_33333417 | 0.03 |
ENSRNOT00000018982
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr8_-_73194837 | 0.02 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr12_-_37444282 | 0.02 |
ENSRNOT00000001368
|
Gtf2h3
|
general transcription factor IIH subunit 3 |
| chr5_+_135574172 | 0.02 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr5_+_160356245 | 0.02 |
ENSRNOT00000085378
|
Casp9
|
caspase 9 |
| chr1_-_101853055 | 0.02 |
ENSRNOT00000084869
|
Grin2d
|
glutamate ionotropic receptor NMDA type subunit 2D |
| chr18_+_44737154 | 0.02 |
ENSRNOT00000021972
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr12_+_37444072 | 0.02 |
ENSRNOT00000077380
ENSRNOT00000092699 ENSRNOT00000001373 ENSRNOT00000092416 |
Eif2b1
|
eukaryotic translation initiation factor 2B subunit 1 alpha |
| chr17_+_77601877 | 0.02 |
ENSRNOT00000091554
ENSRNOT00000024872 |
Prpf18
|
pre-mRNA processing factor 18 |
| chr13_-_95250235 | 0.01 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr15_-_108120279 | 0.01 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr6_-_111477090 | 0.01 |
ENSRNOT00000016389
|
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr10_+_86313383 | 0.01 |
ENSRNOT00000064190
|
Stard3
|
StAR-related lipid transfer domain containing 3 |
| chr5_-_113578928 | 0.01 |
ENSRNOT00000010621
|
Plaa
|
phospholipase A2, activating protein |
| chr8_+_11931767 | 0.00 |
ENSRNOT00000087963
|
Maml2
|
mastermind-like transcriptional coactivator 2 |
| chr6_-_137808303 | 0.00 |
ENSRNOT00000019689
|
Brf1
|
BRF1, RNA polymerase III transcription initiation factor 90 subunit |
| chr7_-_136853154 | 0.00 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr15_+_26887767 | 0.00 |
ENSRNOT00000016674
|
Olr1627
|
olfactory receptor 1627 |
| chr1_-_219532609 | 0.00 |
ENSRNOT00000025641
|
Ankrd13d
|
ankyrin repeat domain 13D |
| chrX_+_6273733 | 0.00 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr2_-_210934749 | 0.00 |
ENSRNOT00000026710
|
Gnai3
|
G protein subunit alpha i3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxg1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000974 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 0.1 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |