Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
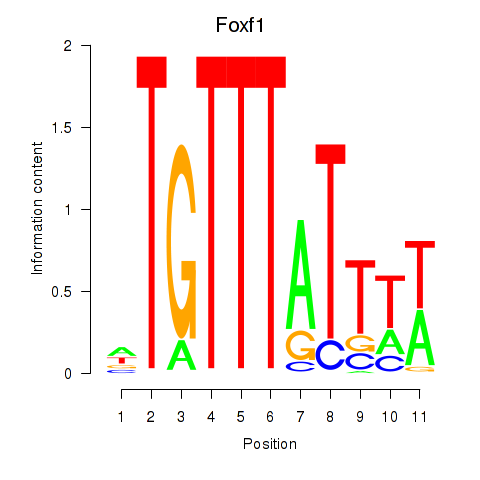
Results for Foxf1
Z-value: 0.47
Transcription factors associated with Foxf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf1
|
ENSRNOG00000049906 | forkhead box F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf1 | rn6_v1_chr19_+_53012332_53012332 | -0.72 | 1.7e-01 | Click! |
Activity profile of Foxf1 motif
Sorted Z-values of Foxf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_77979785 | 0.23 |
ENSRNOT00000084038
|
AC132740.1
|
|
| chr16_+_33373423 | 0.22 |
ENSRNOT00000091672
|
AABR07025316.1
|
|
| chr16_+_33373615 | 0.22 |
ENSRNOT00000079157
|
AABR07025316.1
|
|
| chr1_-_131585731 | 0.19 |
ENSRNOT00000075481
|
AABR07004232.1
|
|
| chr2_-_236823145 | 0.18 |
ENSRNOT00000073001
|
AABR07013425.1
|
|
| chr11_+_69739384 | 0.18 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr2_-_167607919 | 0.17 |
ENSRNOT00000089083
|
AABR07011733.1
|
|
| chr15_+_41937880 | 0.17 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr8_-_115981910 | 0.16 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr5_-_79899054 | 0.15 |
ENSRNOT00000074379
|
AC229945.1
|
|
| chr5_-_7874909 | 0.15 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr14_-_43855745 | 0.15 |
ENSRNOT00000083155
|
AC112624.1
|
|
| chr20_+_7330250 | 0.14 |
ENSRNOT00000090993
|
LOC499407
|
LRRGT00097 |
| chr10_+_53570989 | 0.14 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr8_+_91099491 | 0.13 |
ENSRNOT00000084852
|
Sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr6_-_95502775 | 0.13 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr18_-_55916220 | 0.13 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr20_-_31598118 | 0.13 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr2_-_258997138 | 0.12 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr2_+_127525285 | 0.12 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr1_-_267203986 | 0.10 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr5_+_118574801 | 0.10 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_+_72545596 | 0.09 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr3_-_162059524 | 0.09 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr1_+_259958310 | 0.09 |
ENSRNOT00000019751
|
LOC103689954
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr6_+_135313008 | 0.09 |
ENSRNOT00000030864
|
Tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr4_-_67520356 | 0.09 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr1_-_167911961 | 0.08 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr10_-_74298599 | 0.08 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr3_+_103773459 | 0.08 |
ENSRNOT00000079727
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr1_+_234252757 | 0.08 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr9_+_119542328 | 0.07 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr10_+_46972038 | 0.07 |
ENSRNOT00000023838
|
Mief2
|
mitochondrial elongation factor 2 |
| chr3_+_59981959 | 0.07 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr16_-_81980027 | 0.07 |
ENSRNOT00000043170
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr13_+_83073866 | 0.07 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr17_-_90522091 | 0.07 |
ENSRNOT00000077767
|
Lyst
|
lysosomal trafficking regulator |
| chr18_+_62850588 | 0.07 |
ENSRNOT00000025172
|
Gnal
|
G protein subunit alpha L |
| chr3_-_119330345 | 0.07 |
ENSRNOT00000077398
ENSRNOT00000079191 |
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr3_-_61488696 | 0.07 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr7_+_121930615 | 0.06 |
ENSRNOT00000091270
ENSRNOT00000033975 |
Tnrc6b
|
trinucleotide repeat containing 6B |
| chr15_-_13228607 | 0.06 |
ENSRNOT00000042010
ENSRNOT00000088214 |
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr17_-_21739408 | 0.06 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr13_+_89597138 | 0.06 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr10_+_70262361 | 0.06 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr4_+_10295122 | 0.06 |
ENSRNOT00000017374
|
Ccdc146
|
coiled-coil domain containing 146 |
| chr6_+_122603269 | 0.06 |
ENSRNOT00000005297
|
Spata7
|
spermatogenesis associated 7 |
| chr1_+_8310577 | 0.06 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr13_+_74456487 | 0.06 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr13_+_43850751 | 0.06 |
ENSRNOT00000004995
|
Mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr8_+_50139997 | 0.06 |
ENSRNOT00000022796
|
Bace1
|
beta-secretase 1 |
| chr7_-_118396728 | 0.06 |
ENSRNOT00000066431
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr14_-_108372068 | 0.06 |
ENSRNOT00000088116
ENSRNOT00000091143 ENSRNOT00000085886 |
RGD1305110
|
similar to KIAA1841 protein |
| chr18_+_29966245 | 0.06 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_-_40972826 | 0.06 |
ENSRNOT00000060757
|
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr16_-_69280109 | 0.06 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr7_-_124256077 | 0.06 |
ENSRNOT00000067913
ENSRNOT00000013428 |
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr10_-_27179254 | 0.06 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr4_+_25538497 | 0.06 |
ENSRNOT00000009264
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr8_-_94920441 | 0.05 |
ENSRNOT00000014165
|
Cep162
|
centrosomal protein 162 |
| chr15_-_77736892 | 0.05 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr19_-_11669578 | 0.05 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr2_-_123851854 | 0.05 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr11_-_87924816 | 0.05 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr7_-_119071712 | 0.05 |
ENSRNOT00000037611
|
Myh9l1
|
myosin heavy chain 9-like 1 |
| chr13_+_97838361 | 0.05 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr16_-_6404578 | 0.05 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr14_-_21127868 | 0.05 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr19_+_22450030 | 0.05 |
ENSRNOT00000021739
|
Neto2
|
neuropilin and tolloid like 2 |
| chr8_-_48634797 | 0.05 |
ENSRNOT00000012868
|
Hinfp
|
histone H4 transcription factor |
| chr3_+_48096954 | 0.05 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr8_-_93390305 | 0.05 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr9_-_60923706 | 0.05 |
ENSRNOT00000047517
|
AABR07067749.1
|
|
| chr2_-_96668222 | 0.05 |
ENSRNOT00000016567
|
Pkia
|
cAMP-dependent protein kinase inhibitor alpha |
| chrX_+_43881246 | 0.05 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr4_-_129515435 | 0.05 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr13_-_1946508 | 0.05 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr3_+_41019898 | 0.05 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr2_+_144861455 | 0.04 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr9_-_65693822 | 0.04 |
ENSRNOT00000038431
|
Als2cr12
|
amyotrophic lateral sclerosis 2 chromosome region, candidate 12 |
| chr9_+_118849302 | 0.04 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr4_+_71652354 | 0.04 |
ENSRNOT00000022672
ENSRNOT00000022543 |
Casp2
|
caspase 2 |
| chr3_-_60611924 | 0.04 |
ENSRNOT00000068745
|
Chn1
|
chimerin 1 |
| chr9_+_2190915 | 0.04 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr5_-_60655985 | 0.04 |
ENSRNOT00000093103
|
Fbxo10
|
F-box protein 10 |
| chr1_+_30681681 | 0.04 |
ENSRNOT00000015395
|
Rspo3
|
R-spondin 3 |
| chr14_+_17228856 | 0.04 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr15_-_110612681 | 0.04 |
ENSRNOT00000041551
|
Fgf14
|
fibroblast growth factor 14 |
| chr6_-_76608864 | 0.04 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr8_+_116804451 | 0.04 |
ENSRNOT00000041402
|
Ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr9_-_91100482 | 0.04 |
ENSRNOT00000064642
|
LOC100911860
|
ubiquitin carboxyl-terminal hydrolase 40-like |
| chr10_+_1406486 | 0.04 |
ENSRNOT00000078773
|
LOC679822
|
similar to dynein, cytoplasmic, light peptide |
| chr1_-_253000760 | 0.04 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr14_-_66978499 | 0.04 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr5_+_58636083 | 0.03 |
ENSRNOT00000067281
|
Unc13b
|
unc-13 homolog B |
| chr11_-_45124423 | 0.03 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr8_-_61519507 | 0.03 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr7_+_15785410 | 0.03 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr14_-_21128505 | 0.03 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_+_165805144 | 0.03 |
ENSRNOT00000073716
|
Rabif
|
RAB interacting factor |
| chr3_-_110517163 | 0.03 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr3_-_97993684 | 0.03 |
ENSRNOT00000006481
|
Arl14ep
|
ADP-ribosylation factor like GTPase 14 effector protein |
| chr9_+_66888393 | 0.03 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr1_+_256955652 | 0.03 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr4_+_9981958 | 0.03 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr7_+_11245875 | 0.03 |
ENSRNOT00000027951
|
Gipc3
|
GIPC PDZ domain containing family, member 3 |
| chr3_+_80634470 | 0.03 |
ENSRNOT00000045644
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr17_-_6244612 | 0.03 |
ENSRNOT00000042145
ENSRNOT00000090914 ENSRNOT00000082611 |
Ntrk2
|
neurotrophic receptor tyrosine kinase 2 |
| chr4_+_181428664 | 0.03 |
ENSRNOT00000002518
|
Rep15
|
RAB15 effector protein |
| chr11_-_43099412 | 0.03 |
ENSRNOT00000002281
|
Gabrr3
|
gamma-aminobutyric acid type A receptor rho3 subunit |
| chr5_-_78324278 | 0.03 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr7_-_29378080 | 0.03 |
ENSRNOT00000008317
|
Utp20
|
UTP20 small subunit processome component |
| chr9_-_94495333 | 0.03 |
ENSRNOT00000021507
|
Kcnj13
|
potassium voltage-gated channel subfamily J member 13 |
| chr7_+_142776252 | 0.03 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr8_+_13305152 | 0.03 |
ENSRNOT00000012940
|
Mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr14_-_100184192 | 0.03 |
ENSRNOT00000007044
|
Plek
|
pleckstrin |
| chr18_-_69671199 | 0.03 |
ENSRNOT00000082484
|
Smad4
|
SMAD family member 4 |
| chr2_-_250923744 | 0.03 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr9_-_20195566 | 0.03 |
ENSRNOT00000015223
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr2_+_241909832 | 0.03 |
ENSRNOT00000047975
|
Ppp3ca
|
protein phosphatase 3 catalytic subunit alpha |
| chr6_-_26680284 | 0.03 |
ENSRNOT00000039709
|
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr7_+_142776580 | 0.03 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr8_-_59849427 | 0.03 |
ENSRNOT00000020332
ENSRNOT00000083900 |
Nrg4
|
neuregulin 4 |
| chr18_+_59748444 | 0.03 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr5_-_24560063 | 0.02 |
ENSRNOT00000037869
|
Dpy19l4
|
dpy-19-like 4 (C. elegans) |
| chr20_-_8384088 | 0.02 |
ENSRNOT00000000639
|
Ccdc167
|
coiled-coil domain containing 167 |
| chrX_-_142248369 | 0.02 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr20_+_44060731 | 0.02 |
ENSRNOT00000000737
|
Lama4
|
laminin subunit alpha 4 |
| chr5_-_101006375 | 0.02 |
ENSRNOT00000014008
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr5_+_122100099 | 0.02 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr14_+_8080275 | 0.02 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr2_-_54823917 | 0.02 |
ENSRNOT00000039057
ENSRNOT00000079333 |
Card6
|
caspase recruitment domain family, member 6 |
| chr4_+_181103774 | 0.02 |
ENSRNOT00000084207
ENSRNOT00000055473 ENSRNOT00000077619 |
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr17_-_6660536 | 0.02 |
ENSRNOT00000025812
|
Rmi1
|
RecQ mediated genome instability 1 |
| chr2_+_2903492 | 0.02 |
ENSRNOT00000062017
|
Ttc37
|
tetratricopeptide repeat domain 37 |
| chr10_-_98294522 | 0.02 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr4_+_119171245 | 0.02 |
ENSRNOT00000067651
|
Gkn2
|
gastrokine 2 |
| chr7_-_8060373 | 0.02 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr2_+_69415057 | 0.02 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr10_-_78993045 | 0.02 |
ENSRNOT00000043005
|
LOC100363469
|
ribosomal protein S24-like |
| chr8_-_127871192 | 0.02 |
ENSRNOT00000055928
|
Slc22a14
|
solute carrier family 22, member 14 |
| chr2_+_260444227 | 0.02 |
ENSRNOT00000064249
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr3_-_71845232 | 0.02 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr2_-_250232295 | 0.02 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr10_-_15928169 | 0.02 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr13_+_82479998 | 0.02 |
ENSRNOT00000079872
|
F5
|
coagulation factor V |
| chr10_-_98469799 | 0.02 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr16_-_74710704 | 0.02 |
ENSRNOT00000016859
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chr3_-_51612397 | 0.02 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr1_-_213973163 | 0.02 |
ENSRNOT00000024867
|
LOC100911402
|
cell cycle exit and neuronal differentiation protein 1-like |
| chr4_+_170518673 | 0.02 |
ENSRNOT00000011803
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_-_98197087 | 0.02 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr3_-_13896838 | 0.02 |
ENSRNOT00000025399
|
Fbxw2
|
F-box and WD repeat domain containing 2 |
| chr5_+_128083118 | 0.02 |
ENSRNOT00000079161
|
Zcchc11
|
zinc finger CCHC-type containing 11 |
| chr20_+_33527932 | 0.02 |
ENSRNOT00000066492
|
Nepn
|
nephrocan |
| chrM_+_2740 | 0.02 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr2_-_60657712 | 0.02 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr8_-_26345754 | 0.02 |
ENSRNOT00000091773
|
AC094212.1
|
triosephosphate isomerase |
| chr3_+_10999186 | 0.02 |
ENSRNOT00000037164
|
Nup214
|
nucleoporin 214 |
| chr3_+_129018592 | 0.02 |
ENSRNOT00000007274
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr2_+_60180215 | 0.02 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr6_+_80220547 | 0.01 |
ENSRNOT00000086221
ENSRNOT00000077435 ENSRNOT00000059318 ENSRNOT00000089010 |
Mia2
|
melanoma inhibitory activity 2 |
| chr4_-_8214555 | 0.01 |
ENSRNOT00000084050
ENSRNOT00000076642 |
Kmt2e
|
lysine methyltransferase 2E |
| chr10_+_77762900 | 0.01 |
ENSRNOT00000003308
|
Mmd
|
monocyte to macrophage differentiation-associated |
| chr12_+_703281 | 0.01 |
ENSRNOT00000001464
ENSRNOT00000064951 ENSRNOT00000050733 |
Pds5b
|
PDS5 cohesin associated factor B |
| chr1_+_142834157 | 0.01 |
ENSRNOT00000091569
|
Zfp592
|
zinc finger protein 592 |
| chr7_-_11824742 | 0.01 |
ENSRNOT00000051467
|
Dot1l
|
DOT1 like histone lysine methyltransferase |
| chr19_+_57614628 | 0.01 |
ENSRNOT00000026617
|
Gnpat
|
glyceronephosphate O-acyltransferase |
| chr16_+_20657099 | 0.01 |
ENSRNOT00000027053
|
Kxd1
|
KxDL motif containing 1 |
| chr2_-_157759819 | 0.01 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr7_+_35472102 | 0.01 |
ENSRNOT00000010316
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr11_-_4397361 | 0.01 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr4_+_163293724 | 0.01 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr17_+_1305016 | 0.01 |
ENSRNOT00000025965
|
Ercc6l2
|
ERCC excision repair 6 like 2 |
| chr8_-_128416650 | 0.01 |
ENSRNOT00000046864
|
Scn10a
|
sodium voltage-gated channel alpha subunit 10 |
| chr12_-_40332612 | 0.01 |
ENSRNOT00000001691
|
Atxn2
|
ataxin 2 |
| chr14_+_45033309 | 0.01 |
ENSRNOT00000002933
|
Tlr6
|
toll-like receptor 6 |
| chr7_+_54756043 | 0.01 |
ENSRNOT00000063917
|
Krr1
|
KRR1, small subunit processome component homolog |
| chr5_-_57607725 | 0.01 |
ENSRNOT00000080295
|
Ubap2
|
ubiquitin-associated protein 2 |
| chrX_-_116792864 | 0.01 |
ENSRNOT00000090918
|
Amot
|
angiomotin |
| chr12_-_1195591 | 0.01 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr10_+_105796680 | 0.01 |
ENSRNOT00000000264
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr15_+_87722221 | 0.01 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr10_-_87484935 | 0.01 |
ENSRNOT00000016752
|
Krtap3-1
|
keratin associated protein 3-1 |
| chr1_+_164380577 | 0.01 |
ENSRNOT00000055321
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr18_+_72571327 | 0.01 |
ENSRNOT00000089468
|
Smad2
|
SMAD family member 2 |
| chr15_-_18675431 | 0.01 |
ENSRNOT00000080794
|
Abhd6
|
abhydrolase domain containing 6 |
| chr11_-_28527890 | 0.01 |
ENSRNOT00000002138
|
Cldn8
|
claudin 8 |
| chr17_+_36451058 | 0.01 |
ENSRNOT00000071408
|
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr9_-_15646381 | 0.01 |
ENSRNOT00000040204
|
Mrps10
|
mitochondrial ribosomal protein S10 |
| chr13_-_78521911 | 0.01 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr1_-_72335855 | 0.00 |
ENSRNOT00000021613
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr15_+_36878811 | 0.00 |
ENSRNOT00000090573
|
Rn50_15_0408.6
|
|
| chr2_+_142717281 | 0.00 |
ENSRNOT00000014765
ENSRNOT00000076774 |
Stoml3
|
stomatin like 3 |
| chr4_-_2201749 | 0.00 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr8_+_23193181 | 0.00 |
ENSRNOT00000071703
|
Zfp872
|
zinc finger protein 872 |
| chr2_-_256154584 | 0.00 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr8_+_117347029 | 0.00 |
ENSRNOT00000047717
|
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr3_+_140024043 | 0.00 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr14_-_23604834 | 0.00 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.0 | GO:0090024 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.0 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |