Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Foxd2
Z-value: 0.19
Transcription factors associated with Foxd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd2
|
ENSRNOG00000007759 | forkhead box D2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd2 | rn6_v1_chr5_-_133709712_133709712 | -0.31 | 6.1e-01 | Click! |
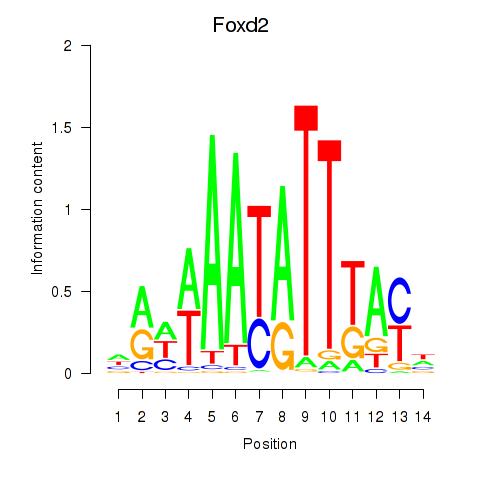
Activity profile of Foxd2 motif
Sorted Z-values of Foxd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_62406873 | 0.07 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr17_+_45199178 | 0.05 |
ENSRNOT00000080047
|
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr13_-_52266100 | 0.05 |
ENSRNOT00000087532
|
AC096239.3
|
|
| chr12_-_51965779 | 0.05 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr5_+_116421894 | 0.04 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr5_+_90818736 | 0.04 |
ENSRNOT00000093480
ENSRNOT00000009083 |
Kdm4c
|
lysine demethylase 4C |
| chr3_-_48831417 | 0.04 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr6_-_125853461 | 0.04 |
ENSRNOT00000007505
|
Atxn3
|
ataxin 3 |
| chr6_-_76535517 | 0.03 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr9_-_30844199 | 0.03 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr3_+_113918629 | 0.03 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr15_-_39742103 | 0.03 |
ENSRNOT00000074587
ENSRNOT00000036781 ENSRNOT00000071919 |
Setdb2
|
SET domain, bifurcated 2 |
| chr9_-_28732919 | 0.03 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr2_+_143475323 | 0.03 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr8_-_13906355 | 0.03 |
ENSRNOT00000029634
|
Cep295
|
centrosomal protein 295 |
| chr4_-_85915099 | 0.03 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chrX_-_142248369 | 0.03 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_+_108301129 | 0.03 |
ENSRNOT00000007993
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr6_-_102472926 | 0.03 |
ENSRNOT00000079351
|
Zfyve26
|
zinc finger FYVE-type containing 26 |
| chr5_+_5616483 | 0.03 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr5_-_83648044 | 0.03 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr5_-_136112344 | 0.03 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr5_+_60850852 | 0.03 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
| chr8_+_82038967 | 0.03 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr10_-_70788309 | 0.03 |
ENSRNOT00000029184
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr15_-_52320385 | 0.03 |
ENSRNOT00000067776
|
Dmtn
|
dematin actin binding protein |
| chr3_-_66885085 | 0.03 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr4_-_168517177 | 0.03 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr8_+_12823155 | 0.03 |
ENSRNOT00000011008
|
Sesn3
|
sestrin 3 |
| chrM_+_11736 | 0.03 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr17_-_51912496 | 0.03 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr19_-_44211208 | 0.03 |
ENSRNOT00000026309
|
Adat1
|
adenosine deaminase, tRNA-specific 1 |
| chr11_+_34865532 | 0.02 |
ENSRNOT00000050342
|
Dyrk1a
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr18_-_77579969 | 0.02 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr18_+_15856801 | 0.02 |
ENSRNOT00000071548
|
Zfp397
|
zinc finger protein 397 |
| chr3_+_9366053 | 0.02 |
ENSRNOT00000073999
|
Fibcd1l1
|
fibrinogen C domain containing 1-like 1 |
| chr14_+_7949239 | 0.02 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr17_-_21705773 | 0.02 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr1_-_164977633 | 0.02 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr8_-_127900463 | 0.02 |
ENSRNOT00000078303
|
Slc22a13
|
solute carrier family 22 member 13 |
| chr17_+_45467015 | 0.02 |
ENSRNOT00000081408
|
Gpx5
|
glutathione peroxidase 5 |
| chr1_+_259958310 | 0.02 |
ENSRNOT00000019751
|
LOC103689954
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr15_-_77736892 | 0.02 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr17_-_84247038 | 0.02 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr20_+_18833481 | 0.02 |
ENSRNOT00000080846
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr2_+_188844073 | 0.02 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr7_+_34326087 | 0.02 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr12_+_7081895 | 0.02 |
ENSRNOT00000047163
|
Hmgb1
|
high mobility group box 1 |
| chr5_+_150833819 | 0.02 |
ENSRNOT00000056170
|
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr8_-_97647072 | 0.02 |
ENSRNOT00000030645
|
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr1_+_190852985 | 0.02 |
ENSRNOT00000040963
|
Polr3e
|
RNA polymerase III subunit E |
| chr13_-_84759098 | 0.02 |
ENSRNOT00000058683
|
LOC685351
|
similar to flavin containing monooxygenase 5 |
| chr10_+_71546977 | 0.02 |
ENSRNOT00000041140
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr17_-_66295014 | 0.02 |
ENSRNOT00000023974
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr17_-_53688966 | 0.02 |
ENSRNOT00000078643
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr10_+_1920529 | 0.02 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr13_-_91776397 | 0.02 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr10_-_37311625 | 0.02 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr14_-_86078954 | 0.02 |
ENSRNOT00000018319
|
Polm
|
DNA polymerase mu |
| chr3_+_47453821 | 0.02 |
ENSRNOT00000081682
|
Tank
|
TRAF family member-associated NFKB activator |
| chr11_-_1983513 | 0.01 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr18_+_30581530 | 0.01 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr6_+_43234526 | 0.01 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr1_+_253221812 | 0.01 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr1_+_189550354 | 0.01 |
ENSRNOT00000083153
|
Exnef
|
exonuclease NEF-sp |
| chr6_-_127620296 | 0.01 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr2_-_186232292 | 0.01 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chr4_-_85174931 | 0.01 |
ENSRNOT00000014324
ENSRNOT00000088085 |
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr2_-_41784929 | 0.01 |
ENSRNOT00000086851
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr15_-_60752106 | 0.01 |
ENSRNOT00000058148
|
Akap11
|
A-kinase anchoring protein 11 |
| chr2_-_250923744 | 0.01 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr2_+_27905535 | 0.01 |
ENSRNOT00000022120
|
Fam169a
|
family with sequence similarity 169, member A |
| chr3_-_163847671 | 0.01 |
ENSRNOT00000076833
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr14_+_85113578 | 0.01 |
ENSRNOT00000011158
|
Nipsnap1
|
nipsnap homolog 1 |
| chr17_-_10622226 | 0.01 |
ENSRNOT00000044559
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr12_-_12782139 | 0.01 |
ENSRNOT00000001392
ENSRNOT00000079836 |
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr14_-_114649173 | 0.01 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_+_67359531 | 0.01 |
ENSRNOT00000013277
ENSRNOT00000082241 |
Adck2
|
aarF domain containing kinase 2 |
| chr6_-_1466201 | 0.01 |
ENSRNOT00000089185
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr8_-_21453190 | 0.01 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr1_+_265809754 | 0.01 |
ENSRNOT00000025221
|
Pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr3_-_12155098 | 0.01 |
ENSRNOT00000082696
|
Garnl3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr2_-_112802073 | 0.01 |
ENSRNOT00000093081
|
Ect2
|
epithelial cell transforming 2 |
| chr5_+_71742911 | 0.01 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr16_-_3765917 | 0.01 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr10_-_89130339 | 0.01 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr14_+_45033309 | 0.01 |
ENSRNOT00000002933
|
Tlr6
|
toll-like receptor 6 |
| chr14_-_2610929 | 0.01 |
ENSRNOT00000036442
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr17_-_57394985 | 0.01 |
ENSRNOT00000019968
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr13_+_27465930 | 0.01 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr15_-_44627765 | 0.01 |
ENSRNOT00000058887
|
Dock5
|
dedicator of cytokinesis 5 |
| chr1_+_141218095 | 0.01 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr5_+_16463597 | 0.01 |
ENSRNOT00000091550
|
AABR07047036.1
|
|
| chr6_-_25507073 | 0.01 |
ENSRNOT00000061176
|
Plb1
|
phospholipase B1 |
| chr3_+_154910626 | 0.01 |
ENSRNOT00000079515
|
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chr7_+_117773948 | 0.01 |
ENSRNOT00000021673
|
Lrrc14
|
leucine rich repeat containing 14 |
| chrX_+_13441558 | 0.01 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr2_+_54466280 | 0.01 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr4_-_29778039 | 0.01 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr7_-_30274984 | 0.01 |
ENSRNOT00000092527
ENSRNOT00000010207 |
Slc17a8
|
solute carrier family 17 member 8 |
| chr13_-_81214494 | 0.01 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr13_+_90260783 | 0.01 |
ENSRNOT00000050547
|
Cd84
|
CD84 molecule |
| chr12_+_47590154 | 0.01 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr19_+_38669230 | 0.01 |
ENSRNOT00000027273
|
Cdh3
|
cadherin 3 |
| chrX_-_143274180 | 0.00 |
ENSRNOT00000051550
|
Mcf2
|
MCF.2 cell line derived transforming sequence |
| chr16_-_48692476 | 0.00 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr19_+_15081590 | 0.00 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr10_+_83154423 | 0.00 |
ENSRNOT00000005939
|
Fam117a
|
family with sequence similarity 117, member A |
| chr13_+_90533365 | 0.00 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr19_+_15081158 | 0.00 |
ENSRNOT00000074070
|
Ces1f
|
carboxylesterase 1F |
| chr14_-_34172893 | 0.00 |
ENSRNOT00000090470
|
Exoc1
|
exocyst complex component 1 |
| chr4_+_35279063 | 0.00 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr5_-_33664435 | 0.00 |
ENSRNOT00000009047
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chrM_-_14061 | 0.00 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_-_253000760 | 0.00 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr4_-_2201749 | 0.00 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr1_-_197821936 | 0.00 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr1_+_185356975 | 0.00 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr10_+_3411380 | 0.00 |
ENSRNOT00000004346
|
RGD1305733
|
similar to RIKEN cDNA 2900011O08 |
| chr1_+_243276403 | 0.00 |
ENSRNOT00000021496
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr3_-_177078594 | 0.00 |
ENSRNOT00000020695
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr11_-_62128044 | 0.00 |
ENSRNOT00000093141
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr8_+_61671513 | 0.00 |
ENSRNOT00000091128
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr14_+_34455934 | 0.00 |
ENSRNOT00000085991
ENSRNOT00000002981 |
Clock
|
clock circadian regulator |
| chrX_-_153878806 | 0.00 |
ENSRNOT00000087990
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr13_+_68707776 | 0.00 |
ENSRNOT00000003558
ENSRNOT00000087997 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr2_-_184309664 | 0.00 |
ENSRNOT00000052051
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr18_-_52017734 | 0.00 |
ENSRNOT00000081020
|
March3
|
membrane associated ring-CH-type finger 3 |
| chr17_-_77527894 | 0.00 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr18_+_3900714 | 0.00 |
ENSRNOT00000092814
|
Lama3
|
laminin subunit alpha 3 |
| chr7_+_73273985 | 0.00 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr3_+_91191837 | 0.00 |
ENSRNOT00000006097
|
Rag2
|
recombination activating 2 |
| chr1_-_275882444 | 0.00 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chrX_+_104684676 | 0.00 |
ENSRNOT00000083229
|
Tnmd
|
tenomodulin |
| chr12_-_8156151 | 0.00 |
ENSRNOT00000064721
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr6_+_107603580 | 0.00 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr7_-_55604403 | 0.00 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr12_+_41486076 | 0.00 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr1_-_205030567 | 0.00 |
ENSRNOT00000023404
|
Ctbp2
|
C-terminal binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |