Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
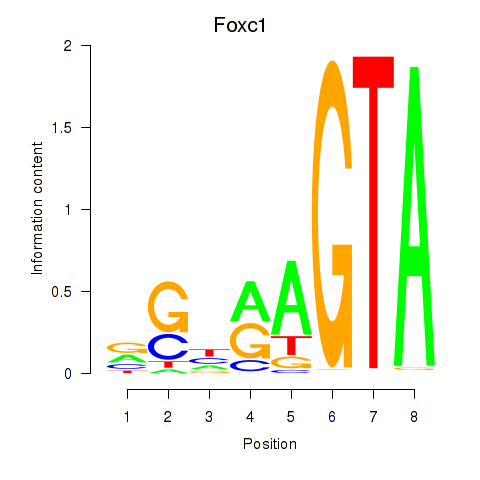
Results for Foxc1
Z-value: 0.36
Transcription factors associated with Foxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc1
|
ENSRNOG00000017800 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc1 | rn6_v1_chr17_-_33951484_33951484 | 0.42 | 4.8e-01 | Click! |
Activity profile of Foxc1 motif
Sorted Z-values of Foxc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_74485139 | 0.13 |
ENSRNOT00000022598
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr5_+_118574801 | 0.12 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr5_-_105579959 | 0.10 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr5_+_76092287 | 0.10 |
ENSRNOT00000020207
|
Zfp483
|
zinc finger protein 483 |
| chr2_-_149444548 | 0.10 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr14_+_71533063 | 0.09 |
ENSRNOT00000004231
|
Prom1
|
prominin 1 |
| chr7_-_49250953 | 0.08 |
ENSRNOT00000066975
ENSRNOT00000082141 |
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr12_-_5773036 | 0.08 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr17_-_77261731 | 0.08 |
ENSRNOT00000066850
|
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr2_+_95077577 | 0.08 |
ENSRNOT00000046368
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr7_-_113937941 | 0.08 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr18_+_56379890 | 0.08 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr4_+_158243086 | 0.08 |
ENSRNOT00000032112
|
Ano2
|
anoctamin 2 |
| chr9_-_94495333 | 0.06 |
ENSRNOT00000021507
|
Kcnj13
|
potassium voltage-gated channel subfamily J member 13 |
| chr15_-_83494423 | 0.06 |
ENSRNOT00000037588
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr6_+_99402360 | 0.06 |
ENSRNOT00000078498
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr3_+_79823945 | 0.06 |
ENSRNOT00000014484
|
Celf1
|
CUGBP, Elav-like family member 1 |
| chr17_+_76532611 | 0.06 |
ENSRNOT00000024099
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr6_+_64252970 | 0.06 |
ENSRNOT00000093700
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr1_-_236729412 | 0.06 |
ENSRNOT00000054794
|
Prune2
|
prune homolog 2 |
| chr10_-_38969501 | 0.06 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr13_-_103080920 | 0.06 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr3_+_60024013 | 0.05 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr3_-_123849788 | 0.05 |
ENSRNOT00000028869
|
Rnf24
|
ring finger protein 24 |
| chr13_-_35668968 | 0.05 |
ENSRNOT00000042862
ENSRNOT00000003428 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr1_+_27476375 | 0.05 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr1_+_141120166 | 0.05 |
ENSRNOT00000050759
|
Fanci
|
Fanconi anemia, complementation group I |
| chr7_+_112057030 | 0.04 |
ENSRNOT00000035216
|
AABR07058366.1
|
|
| chr15_-_61648267 | 0.04 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr2_-_258997138 | 0.04 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr6_+_64297888 | 0.04 |
ENSRNOT00000050222
ENSRNOT00000083088 ENSRNOT00000093147 |
Nrcam
|
neuronal cell adhesion molecule |
| chr3_+_176865156 | 0.04 |
ENSRNOT00000019084
|
Zgpat
|
zinc finger CCCH-type and G-patch domain containing |
| chr2_+_84678948 | 0.04 |
ENSRNOT00000046325
|
Fam173b
|
family with sequence similarity 173, member B |
| chr3_-_162579201 | 0.04 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr3_+_62648447 | 0.04 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr4_-_165456677 | 0.04 |
ENSRNOT00000082207
|
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr14_-_5428757 | 0.04 |
ENSRNOT00000002897
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr1_+_215610368 | 0.04 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chr17_-_53688966 | 0.04 |
ENSRNOT00000078643
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr11_+_44001579 | 0.03 |
ENSRNOT00000002258
|
Gpr15
|
G protein-coupled receptor 15 |
| chrX_+_88298266 | 0.03 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr7_+_130308532 | 0.03 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr8_-_49308806 | 0.03 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr2_-_30458542 | 0.03 |
ENSRNOT00000072926
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr6_+_106052212 | 0.03 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr11_-_64952687 | 0.03 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr13_-_70980913 | 0.03 |
ENSRNOT00000003713
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr16_-_79973735 | 0.03 |
ENSRNOT00000057845
ENSRNOT00000086896 |
Dlgap2
|
DLG associated protein 2 |
| chr1_-_44615760 | 0.03 |
ENSRNOT00000022148
|
Nox3
|
NADPH oxidase 3 |
| chr16_-_32439421 | 0.03 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chr3_-_112876773 | 0.03 |
ENSRNOT00000015086
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr3_+_8802852 | 0.03 |
ENSRNOT00000033934
|
Lrrc8a
|
leucine rich repeat containing 8 family, member A |
| chr2_-_178616719 | 0.03 |
ENSRNOT00000078610
|
Tmem144
|
transmembrane protein 144 |
| chr4_-_66955732 | 0.03 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr8_+_132828091 | 0.02 |
ENSRNOT00000008269
|
Ccr9
|
C-C motif chemokine receptor 9 |
| chr13_-_111972603 | 0.02 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr11_+_61748883 | 0.02 |
ENSRNOT00000093552
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr1_-_81127059 | 0.02 |
ENSRNOT00000026252
|
Zfp94
|
zinc finger protein 94 |
| chr1_+_154131926 | 0.02 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr1_-_253000760 | 0.02 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr5_-_50138475 | 0.02 |
ENSRNOT00000011969
|
Slc35a1
|
solute carrier family 35 member A1 |
| chr1_+_88885937 | 0.02 |
ENSRNOT00000029719
|
Nfkbid
|
NFKB inhibitor delta |
| chr1_+_215609645 | 0.02 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr9_-_81772851 | 0.02 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr10_-_84698886 | 0.02 |
ENSRNOT00000067542
|
Nfe2l1
|
nuclear factor, erythroid 2-like 1 |
| chr14_+_81858737 | 0.02 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr20_+_2501252 | 0.02 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr16_+_35573058 | 0.02 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr19_+_52086325 | 0.02 |
ENSRNOT00000020341
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr5_+_117583502 | 0.02 |
ENSRNOT00000010933
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr3_+_172719432 | 0.02 |
ENSRNOT00000033863
|
Zfp831
|
zinc finger protein 831 |
| chr3_-_160301552 | 0.02 |
ENSRNOT00000014498
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr3_+_122544788 | 0.02 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr3_+_128756799 | 0.02 |
ENSRNOT00000049855
ENSRNOT00000042853 |
Plcb4
|
phospholipase C, beta 4 |
| chr8_-_103190243 | 0.02 |
ENSRNOT00000075305
|
Chst2
|
carbohydrate sulfotransferase 2 |
| chr6_-_26875376 | 0.02 |
ENSRNOT00000011838
|
Tmem214
|
transmembrane protein 214 |
| chr10_-_91986632 | 0.02 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr13_-_86671515 | 0.02 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr11_+_28692708 | 0.02 |
ENSRNOT00000002135
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr8_-_109560747 | 0.02 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr9_-_92405117 | 0.02 |
ENSRNOT00000079452
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr8_+_79489790 | 0.02 |
ENSRNOT00000091722
|
Prtg
|
protogenin |
| chr20_-_45126062 | 0.02 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr1_-_261669584 | 0.02 |
ENSRNOT00000020568
ENSRNOT00000076555 |
Crtac1
|
cartilage acidic protein 1 |
| chrX_+_71342775 | 0.02 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr16_+_16949232 | 0.01 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr8_+_57936650 | 0.01 |
ENSRNOT00000089686
|
Exph5
|
exophilin 5 |
| chr18_+_30375798 | 0.01 |
ENSRNOT00000060497
|
Pcdhb2
|
protocadherin beta 2 |
| chr5_-_75116490 | 0.01 |
ENSRNOT00000042788
|
Txndc8
|
thioredoxin domain containing 8 |
| chr8_+_59457018 | 0.01 |
ENSRNOT00000017900
|
Ireb2
|
iron responsive element binding protein 2 |
| chr1_-_175895510 | 0.01 |
ENSRNOT00000064535
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr3_+_94035905 | 0.01 |
ENSRNOT00000014767
|
RGD1563222
|
similar to RIKEN cDNA A930018P22 |
| chr15_+_27739251 | 0.01 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr10_+_11786121 | 0.01 |
ENSRNOT00000033919
|
Slx4
|
SLX4 structure-specific endonuclease subunit |
| chr2_+_60131776 | 0.01 |
ENSRNOT00000080786
|
Prlr
|
prolactin receptor |
| chr7_+_138707426 | 0.01 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chr15_-_37926715 | 0.01 |
ENSRNOT00000013901
|
Xpo4
|
exportin 4 |
| chr3_+_80468154 | 0.01 |
ENSRNOT00000021837
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr19_+_39063998 | 0.01 |
ENSRNOT00000081116
|
Has3
|
hyaluronan synthase 3 |
| chr5_-_77031120 | 0.01 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr9_-_11108741 | 0.01 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr10_-_78690857 | 0.01 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr10_+_53713938 | 0.01 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr18_+_29951094 | 0.01 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr3_+_80556668 | 0.01 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr14_+_11199404 | 0.01 |
ENSRNOT00000003106
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr8_+_22423467 | 0.01 |
ENSRNOT00000085320
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr15_+_105851542 | 0.01 |
ENSRNOT00000086959
|
Rap2a
|
RAS related protein 2a |
| chr10_-_98544447 | 0.01 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr10_+_53781239 | 0.01 |
ENSRNOT00000082871
|
Myh2
|
myosin heavy chain 2 |
| chr16_+_26906716 | 0.01 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr15_+_32809069 | 0.00 |
ENSRNOT00000070848
|
AABR07017902.1
|
|
| chr7_-_117068332 | 0.00 |
ENSRNOT00000082433
|
Fam83h
|
family with sequence similarity 83, member H |
| chr12_-_50314406 | 0.00 |
ENSRNOT00000079256
|
Hps4
|
Hermansky-Pudlak syndrome 4 |
| chr7_+_13996966 | 0.00 |
ENSRNOT00000009615
|
Olr1087
|
olfactory receptor 1087 |
| chr9_+_100137881 | 0.00 |
ENSRNOT00000036328
|
Gpr35
|
G protein-coupled receptor 35 |
| chr14_+_37116492 | 0.00 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chrX_+_15598652 | 0.00 |
ENSRNOT00000082383
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr1_-_174119815 | 0.00 |
ENSRNOT00000019368
|
Trim66
|
tripartite motif-containing 66 |
| chr14_-_14689554 | 0.00 |
ENSRNOT00000002814
|
Fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr18_-_35817117 | 0.00 |
ENSRNOT00000015172
ENSRNOT00000085714 |
Mcc
|
mutated in colorectal cancers |
| chr18_+_2416871 | 0.00 |
ENSRNOT00000081399
|
Gata6
|
GATA binding protein 6 |
| chr1_+_44311513 | 0.00 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr15_-_29993639 | 0.00 |
ENSRNOT00000071822
|
AABR07017677.1
|
|
| chr1_+_125367280 | 0.00 |
ENSRNOT00000022049
|
Apba2
|
amyloid beta precursor protein binding family A member 2 |
| chr9_-_71798265 | 0.00 |
ENSRNOT00000043766
|
Crygb
|
crystallin, gamma B |
| chr3_+_170380719 | 0.00 |
ENSRNOT00000055101
|
Cstf1
|
cleavage stimulation factor subunit 1 |
| chr10_-_5344121 | 0.00 |
ENSRNOT00000003479
|
Nubp1
|
nucleotide binding protein 1 |
| chr1_-_66645214 | 0.00 |
ENSRNOT00000084168
|
LOC100365062
|
eukaryotic translation initiation factor 3, subunit 6 48kDa-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0038086 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |