Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
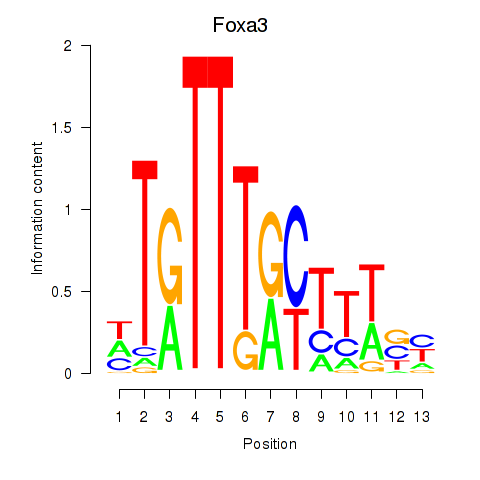
Results for Foxa3
Z-value: 0.42
Transcription factors associated with Foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxa3
|
ENSRNOG00000014256 | forkhead box A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxa3 | rn6_v1_chr1_-_79930263_79930263 | -0.66 | 2.3e-01 | Click! |
Activity profile of Foxa3 motif
Sorted Z-values of Foxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_51614676 | 0.22 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr12_-_36398206 | 0.22 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr9_+_27034022 | 0.20 |
ENSRNOT00000029696
|
Paqr8
|
progestin and adipoQ receptor family member 8 |
| chr10_-_27179900 | 0.18 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr3_-_158328881 | 0.17 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr1_-_78521999 | 0.17 |
ENSRNOT00000021223
|
Arhgap35
|
Rho GTPase activating protein 35 |
| chr10_-_37311625 | 0.16 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr7_+_23403891 | 0.15 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr13_+_43850751 | 0.14 |
ENSRNOT00000004995
|
Mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr18_+_30890869 | 0.14 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr14_+_63405408 | 0.14 |
ENSRNOT00000086658
|
AABR07015559.1
|
|
| chr3_+_131351587 | 0.14 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr3_-_119330345 | 0.13 |
ENSRNOT00000077398
ENSRNOT00000079191 |
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr9_+_118849302 | 0.13 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr6_+_135313008 | 0.13 |
ENSRNOT00000030864
|
Tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr20_-_6251634 | 0.12 |
ENSRNOT00000059194
|
Stk38
|
serine/threonine kinase 38 |
| chr7_+_71157664 | 0.12 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr10_-_27179254 | 0.12 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr8_-_115981910 | 0.12 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr12_-_40822159 | 0.12 |
ENSRNOT00000001826
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr2_-_258997138 | 0.11 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr1_-_267203986 | 0.11 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr1_+_214562897 | 0.10 |
ENSRNOT00000085125
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr1_+_156552328 | 0.10 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr16_-_81980027 | 0.09 |
ENSRNOT00000043170
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr5_+_165415136 | 0.09 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr2_-_111065413 | 0.09 |
ENSRNOT00000041111
|
Nlgn1
|
neuroligin 1 |
| chr3_-_71845232 | 0.09 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr6_-_104631355 | 0.09 |
ENSRNOT00000007825
|
Slc10a1
|
solute carrier family 10 member 1 |
| chr7_-_120770435 | 0.08 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr8_-_102149912 | 0.08 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr10_+_11393103 | 0.08 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr4_+_170518673 | 0.08 |
ENSRNOT00000011803
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_+_77107536 | 0.07 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chr3_-_162059524 | 0.07 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr10_-_103687425 | 0.07 |
ENSRNOT00000039284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr10_+_86399827 | 0.07 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr6_-_67084234 | 0.07 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr12_-_1195591 | 0.06 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr1_+_80141630 | 0.06 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr16_-_25192675 | 0.06 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chr5_+_58359498 | 0.06 |
ENSRNOT00000000141
|
Phf24
|
PHD finger protein 24 |
| chr2_+_44289393 | 0.06 |
ENSRNOT00000018877
|
Il6st
|
interleukin 6 signal transducer |
| chr11_-_45124423 | 0.06 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr4_+_181103774 | 0.05 |
ENSRNOT00000084207
ENSRNOT00000055473 ENSRNOT00000077619 |
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr20_-_27308069 | 0.05 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr1_-_131460473 | 0.05 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_+_17228856 | 0.05 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr20_-_38985036 | 0.05 |
ENSRNOT00000001066
|
Serinc1
|
serine incorporator 1 |
| chr9_+_66888393 | 0.05 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr1_+_277459200 | 0.05 |
ENSRNOT00000086008
|
AABR07007023.1
|
|
| chr15_-_28406046 | 0.04 |
ENSRNOT00000015418
|
Zfp219
|
zinc finger protein 219 |
| chr14_+_39964588 | 0.04 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr13_-_84619023 | 0.04 |
ENSRNOT00000089827
ENSRNOT00000045056 |
Pogk
|
pogo transposable element with KRAB domain |
| chr14_-_21128505 | 0.04 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr7_-_138483612 | 0.04 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr6_+_93462852 | 0.04 |
ENSRNOT00000089473
|
Arid4a
|
AT-rich interaction domain 4A |
| chr8_-_94920441 | 0.04 |
ENSRNOT00000014165
|
Cep162
|
centrosomal protein 162 |
| chr13_+_83073866 | 0.04 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr6_+_50528823 | 0.04 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr1_+_234252757 | 0.04 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr15_+_34216833 | 0.04 |
ENSRNOT00000025260
|
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr6_-_108167185 | 0.04 |
ENSRNOT00000015545
|
Aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr11_-_70498391 | 0.04 |
ENSRNOT00000040167
ENSRNOT00000086495 |
Slc12a8
|
solute carrier family 12, member 8 |
| chr2_-_27364906 | 0.04 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr10_-_88060561 | 0.04 |
ENSRNOT00000019133
|
Krt19
|
keratin 19 |
| chr14_+_8080275 | 0.04 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr1_-_40921508 | 0.03 |
ENSRNOT00000026433
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr2_+_34546988 | 0.03 |
ENSRNOT00000072577
|
Adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr6_-_108660063 | 0.03 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr3_+_122836446 | 0.03 |
ENSRNOT00000028816
|
Ebf4
|
early B-cell factor 4 |
| chr13_-_50509916 | 0.03 |
ENSRNOT00000076747
|
Ren
|
renin |
| chrX_+_35599258 | 0.03 |
ENSRNOT00000072627
ENSRNOT00000005061 |
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr18_+_16146447 | 0.03 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr11_-_87924816 | 0.03 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr20_+_7330250 | 0.03 |
ENSRNOT00000090993
|
LOC499407
|
LRRGT00097 |
| chr18_+_59748444 | 0.03 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr7_+_63467216 | 0.03 |
ENSRNOT00000064349
|
LOC100909505
|
N-acetylglucosamine-6-sulfatase-like |
| chr18_-_58097316 | 0.03 |
ENSRNOT00000088527
|
AABR07032265.1
|
|
| chr14_-_21127868 | 0.03 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr13_-_91872954 | 0.03 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr13_+_74456487 | 0.03 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr1_+_264741911 | 0.03 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr14_+_42714315 | 0.03 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr6_+_108167716 | 0.03 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr3_-_90751055 | 0.03 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr7_+_26522314 | 0.03 |
ENSRNOT00000011572
|
Slc41a2
|
solute carrier family 41 member 2 |
| chr6_+_42947334 | 0.03 |
ENSRNOT00000041376
|
RGD1563157
|
similar to 60S ribosomal protein L35 |
| chr2_+_69415057 | 0.03 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr6_+_86872903 | 0.02 |
ENSRNOT00000086894
|
Fancm
|
Fanconi anemia, complementation group M |
| chr7_-_15072703 | 0.02 |
ENSRNOT00000087294
|
Zfp799
|
zinc finger protein 799 |
| chr8_+_85417838 | 0.02 |
ENSRNOT00000012257
|
Ick
|
intestinal cell kinase |
| chr11_+_74984613 | 0.02 |
ENSRNOT00000035049
|
Atp13a5
|
ATPase 13A5 |
| chr16_-_20994379 | 0.02 |
ENSRNOT00000027639
|
Tmem161a
|
transmembrane protein 161A |
| chr13_-_74331214 | 0.02 |
ENSRNOT00000006018
|
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr2_-_23909016 | 0.02 |
ENSRNOT00000084044
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr3_+_148635775 | 0.02 |
ENSRNOT00000067261
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr1_-_220165678 | 0.02 |
ENSRNOT00000026915
|
Bbs1
|
Bardet-Biedl syndrome 1 |
| chr7_-_97605113 | 0.02 |
ENSRNOT00000089734
|
AABR07058000.1
|
|
| chr20_+_29655226 | 0.02 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr2_-_147819335 | 0.02 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr20_+_44060731 | 0.02 |
ENSRNOT00000000737
|
Lama4
|
laminin subunit alpha 4 |
| chr7_+_132857628 | 0.02 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr19_-_11669578 | 0.02 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr8_-_90483077 | 0.02 |
ENSRNOT00000011864
|
Phip
|
pleckstrin homology domain interacting protein |
| chr11_-_782954 | 0.02 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr5_+_120568242 | 0.02 |
ENSRNOT00000050597
|
Lepr
|
leptin receptor |
| chr1_-_189182306 | 0.02 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr5_+_49311030 | 0.02 |
ENSRNOT00000010850
|
Cnr1
|
cannabinoid receptor 1 |
| chr9_-_17206994 | 0.02 |
ENSRNOT00000026195
|
Gtpbp2
|
GTP binding protein 2 |
| chr9_+_111649631 | 0.01 |
ENSRNOT00000056430
|
Fer
|
FER tyrosine kinase |
| chrX_+_113510621 | 0.01 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_144861455 | 0.01 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr1_-_134870255 | 0.01 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_-_46404642 | 0.01 |
ENSRNOT00000083698
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr2_+_197752544 | 0.01 |
ENSRNOT00000079196
ENSRNOT00000075511 |
Ensa
|
endosulfine alpha |
| chr11_-_69223158 | 0.01 |
ENSRNOT00000067060
|
Mylk
|
myosin light chain kinase |
| chr10_-_55783489 | 0.01 |
ENSRNOT00000010415
|
Alox15b
|
arachidonate 15-lipoxygenase, type B |
| chr1_+_72545596 | 0.01 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr7_-_15073052 | 0.01 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr18_+_3887419 | 0.01 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr11_-_15858281 | 0.01 |
ENSRNOT00000090586
|
AABR07033287.1
|
|
| chr8_+_91099491 | 0.01 |
ENSRNOT00000084852
|
Sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr1_+_178636544 | 0.01 |
ENSRNOT00000092099
|
Spon1
|
spondin 1 |
| chr1_+_107262659 | 0.01 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr5_+_61657507 | 0.00 |
ENSRNOT00000013228
|
Tmod1
|
tropomodulin 1 |
| chr1_-_90520012 | 0.00 |
ENSRNOT00000028698
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr3_+_137154086 | 0.00 |
ENSRNOT00000034252
|
Otor
|
otoraplin |
| chr13_-_1946508 | 0.00 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr16_-_3765917 | 0.00 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr10_-_87521514 | 0.00 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr2_+_154555871 | 0.00 |
ENSRNOT00000076292
|
Gmps
|
guanine monophosphate synthase |
| chr9_-_55256340 | 0.00 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr4_+_62220736 | 0.00 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:2000426 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.0 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.0 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.0 | GO:1990743 | protein sialylation(GO:1990743) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070110 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |