Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
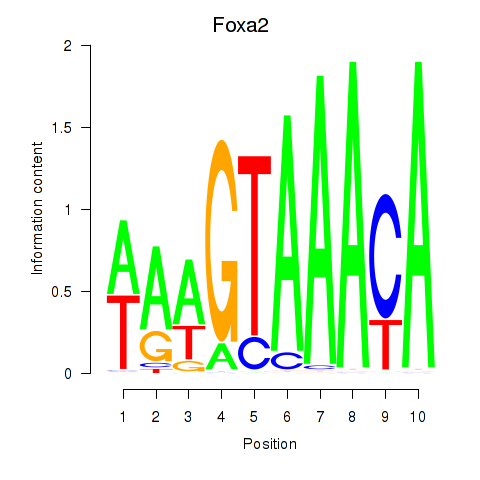
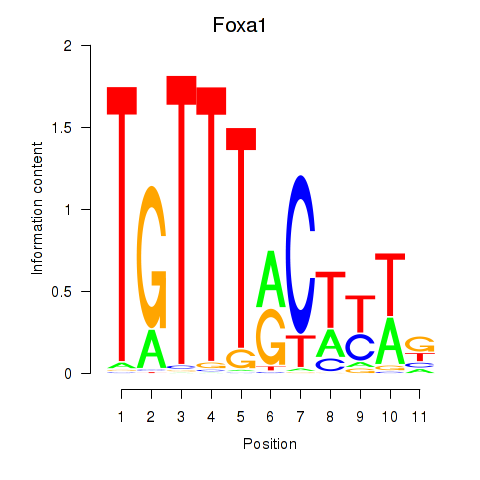
Results for Foxa2_Foxa1
Z-value: 0.36
Transcription factors associated with Foxa2_Foxa1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxa2
|
ENSRNOG00000013133 | forkhead box A2 |
|
Foxa1
|
ENSRNOG00000009284 | forkhead box A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxa1 | rn6_v1_chr6_-_78549669_78549669 | 0.83 | 8.3e-02 | Click! |
Activity profile of Foxa2_Foxa1 motif
Sorted Z-values of Foxa2_Foxa1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_55916220 | 0.38 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr8_+_40009691 | 0.23 |
ENSRNOT00000042679
|
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr9_-_15274917 | 0.22 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr3_-_158328881 | 0.19 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr9_+_27034022 | 0.17 |
ENSRNOT00000029696
|
Paqr8
|
progestin and adipoQ receptor family member 8 |
| chr1_-_267203986 | 0.17 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr9_+_118849302 | 0.16 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr9_+_119542328 | 0.16 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr7_+_41475163 | 0.15 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr12_-_36398206 | 0.14 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr10_-_37311625 | 0.13 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr10_-_103687425 | 0.13 |
ENSRNOT00000039284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr9_+_47281961 | 0.12 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr20_+_29655226 | 0.12 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr10_+_11393103 | 0.11 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr1_-_131460473 | 0.10 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_27476375 | 0.10 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr11_-_45124423 | 0.10 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr10_-_88060561 | 0.09 |
ENSRNOT00000019133
|
Krt19
|
keratin 19 |
| chr1_+_261337594 | 0.09 |
ENSRNOT00000019874
|
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr13_+_43850751 | 0.09 |
ENSRNOT00000004995
|
Mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr11_-_11585078 | 0.09 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr3_+_162357356 | 0.09 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr7_+_99954492 | 0.08 |
ENSRNOT00000005885
|
Trib1
|
tribbles pseudokinase 1 |
| chr1_-_134870255 | 0.08 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr1_+_248228496 | 0.08 |
ENSRNOT00000015406
|
Uhrf2
|
ubiquitin like with PHD and ring finger domains 2 |
| chr12_-_40822159 | 0.07 |
ENSRNOT00000001826
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr3_-_141411170 | 0.07 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr7_+_58814805 | 0.07 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr7_-_138483612 | 0.07 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr15_+_23792931 | 0.07 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr6_+_42947334 | 0.06 |
ENSRNOT00000041376
|
RGD1563157
|
similar to 60S ribosomal protein L35 |
| chrX_+_35599258 | 0.06 |
ENSRNOT00000072627
ENSRNOT00000005061 |
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr11_+_65743892 | 0.06 |
ENSRNOT00000085226
|
AC106292.2
|
|
| chr8_-_115981910 | 0.06 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr2_-_258997138 | 0.06 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr1_+_156552328 | 0.05 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr6_+_106052212 | 0.05 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr2_+_200572502 | 0.05 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr1_+_81779380 | 0.05 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chr13_-_90710148 | 0.05 |
ENSRNOT00000010113
ENSRNOT00000080638 |
Kcnj9
|
potassium voltage-gated channel subfamily J member 9 |
| chr2_+_200603426 | 0.05 |
ENSRNOT00000072189
|
LOC102550892
|
zinc finger protein 697-like |
| chr10_+_105393072 | 0.05 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chr4_-_15505362 | 0.05 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr10_+_58860940 | 0.05 |
ENSRNOT00000056551
ENSRNOT00000074523 |
XAF1
|
XIAP associated factor-1 |
| chr7_-_97605113 | 0.05 |
ENSRNOT00000089734
|
AABR07058000.1
|
|
| chr5_+_165415136 | 0.04 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr7_-_122329443 | 0.04 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr8_-_104593625 | 0.04 |
ENSRNOT00000016625
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr7_-_28040510 | 0.04 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr1_-_90520344 | 0.04 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr10_-_51669297 | 0.04 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr16_-_81980027 | 0.04 |
ENSRNOT00000043170
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr9_+_67234303 | 0.04 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr10_-_27179254 | 0.04 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr10_-_15928169 | 0.04 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr3_-_162059524 | 0.04 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr14_+_42015347 | 0.04 |
ENSRNOT00000044017
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr10_+_77762900 | 0.04 |
ENSRNOT00000003308
|
Mmd
|
monocyte to macrophage differentiation-associated |
| chr2_+_54897424 | 0.04 |
ENSRNOT00000017665
|
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr5_+_48290061 | 0.04 |
ENSRNOT00000076508
|
Ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr13_-_94859390 | 0.04 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr16_+_50152008 | 0.04 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr3_+_63379031 | 0.04 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr4_-_68819872 | 0.04 |
ENSRNOT00000033265
ENSRNOT00000081884 |
Clec5a
|
C-type lectin domain family 5, member A |
| chr9_+_18564927 | 0.04 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr2_-_250232295 | 0.04 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr10_+_81913689 | 0.04 |
ENSRNOT00000003780
|
Tob1
|
transducer of ErbB-2.1 |
| chr13_+_83073866 | 0.04 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr6_-_50786967 | 0.04 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr17_+_81922329 | 0.04 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr6_+_93462852 | 0.03 |
ENSRNOT00000089473
|
Arid4a
|
AT-rich interaction domain 4A |
| chr17_-_1093873 | 0.03 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr3_-_60795951 | 0.03 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr5_+_59178846 | 0.03 |
ENSRNOT00000021227
|
Tmem8b
|
transmembrane protein 8B |
| chr5_+_116420690 | 0.03 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr10_+_80953006 | 0.03 |
ENSRNOT00000035567
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr9_+_60039297 | 0.03 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr4_-_81241152 | 0.03 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr20_+_5646097 | 0.03 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr1_-_236729412 | 0.03 |
ENSRNOT00000054794
|
Prune2
|
prune homolog 2 |
| chr4_-_55011415 | 0.03 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr9_-_46475040 | 0.03 |
ENSRNOT00000078196
|
Rfx8
|
RFX family member 8, lacking RFX DNA binding domain |
| chr10_+_17486579 | 0.03 |
ENSRNOT00000080404
|
Stk10
|
serine/threonine kinase 10 |
| chr2_+_44289393 | 0.03 |
ENSRNOT00000018877
|
Il6st
|
interleukin 6 signal transducer |
| chr15_+_55126953 | 0.03 |
ENSRNOT00000021430
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr3_-_82856171 | 0.03 |
ENSRNOT00000088555
|
RGD1564664
|
similar to LOC387763 protein |
| chr9_-_63641400 | 0.03 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr15_+_34520142 | 0.03 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr2_+_198359754 | 0.03 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr2_-_192671059 | 0.03 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr2_+_104744461 | 0.03 |
ENSRNOT00000016083
ENSRNOT00000082627 |
Cp
|
ceruloplasmin |
| chr9_+_71915421 | 0.03 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr7_+_28414350 | 0.03 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr8_+_123126667 | 0.03 |
ENSRNOT00000015643
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr16_-_19583386 | 0.03 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr17_-_84830185 | 0.03 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chr14_-_44767120 | 0.03 |
ENSRNOT00000003991
|
Wdr19
|
WD repeat domain 19 |
| chr7_+_54247460 | 0.03 |
ENSRNOT00000005361
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr6_+_27887797 | 0.03 |
ENSRNOT00000015827
|
Asxl2
|
additional sex combs like 2, transcriptional regulator |
| chr15_+_33074441 | 0.03 |
ENSRNOT00000075610
|
Mmp14
|
matrix metallopeptidase 14 |
| chr6_-_3355339 | 0.03 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr3_+_131351587 | 0.02 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr1_-_90520012 | 0.02 |
ENSRNOT00000028698
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr4_+_168599331 | 0.02 |
ENSRNOT00000086719
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr10_-_95212111 | 0.02 |
ENSRNOT00000020795
|
Kpna2
|
karyopherin subunit alpha 2 |
| chr10_-_57064600 | 0.02 |
ENSRNOT00000032926
|
Cxcl16
|
C-X-C motif chemokine ligand 16 |
| chr5_+_25168295 | 0.02 |
ENSRNOT00000020245
|
Fsbp
|
fibrinogen silencer binding protein |
| chr6_-_104631355 | 0.02 |
ENSRNOT00000007825
|
Slc10a1
|
solute carrier family 10 member 1 |
| chr15_-_77736892 | 0.02 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr13_+_97838361 | 0.02 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr2_+_234375315 | 0.02 |
ENSRNOT00000071270
|
LOC102549542
|
elongation of very long chain fatty acids protein 6-like |
| chr4_-_84131030 | 0.02 |
ENSRNOT00000012196
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chrX_+_62754634 | 0.02 |
ENSRNOT00000016669
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_+_72545596 | 0.02 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr9_-_17206994 | 0.02 |
ENSRNOT00000026195
|
Gtpbp2
|
GTP binding protein 2 |
| chr3_-_150855228 | 0.02 |
ENSRNOT00000089935
|
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr9_+_111220858 | 0.02 |
ENSRNOT00000076669
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr2_+_236233239 | 0.02 |
ENSRNOT00000013694
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr1_+_82452469 | 0.02 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr5_-_88629491 | 0.02 |
ENSRNOT00000058906
|
Tle1
|
transducin like enhancer of split 1 |
| chr13_-_79744939 | 0.02 |
ENSRNOT00000076706
|
Suco
|
SUN domain containing ossification factor |
| chr13_-_84619023 | 0.02 |
ENSRNOT00000089827
ENSRNOT00000045056 |
Pogk
|
pogo transposable element with KRAB domain |
| chr6_+_58468155 | 0.02 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr10_+_18996523 | 0.02 |
ENSRNOT00000046135
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr5_+_157165341 | 0.02 |
ENSRNOT00000087072
|
Pla2g2c
|
phospholipase A2, group IIC |
| chr13_-_74331214 | 0.02 |
ENSRNOT00000006018
|
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr3_+_120726906 | 0.02 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr9_+_111028575 | 0.02 |
ENSRNOT00000043451
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_78042859 | 0.02 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr5_+_113725717 | 0.02 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr7_+_120176530 | 0.02 |
ENSRNOT00000087548
|
Triobp
|
TRIO and F-actin binding protein |
| chr2_-_183128932 | 0.02 |
ENSRNOT00000031179
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr2_+_69415057 | 0.02 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr1_+_220325352 | 0.02 |
ENSRNOT00000027258
|
LOC108348098
|
breast cancer metastasis-suppressor 1 homolog |
| chr10_-_27179900 | 0.02 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr2_-_54823917 | 0.02 |
ENSRNOT00000039057
ENSRNOT00000079333 |
Card6
|
caspase recruitment domain family, member 6 |
| chr15_+_24942218 | 0.02 |
ENSRNOT00000016629
|
Peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr13_+_47602692 | 0.02 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr6_+_22696397 | 0.01 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chr11_-_70498391 | 0.01 |
ENSRNOT00000040167
ENSRNOT00000086495 |
Slc12a8
|
solute carrier family 12, member 8 |
| chr8_-_130127392 | 0.01 |
ENSRNOT00000026159
|
Cck
|
cholecystokinin |
| chr8_+_50525091 | 0.01 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr7_+_91833297 | 0.01 |
ENSRNOT00000079792
|
Slc30a8
|
solute carrier family 30 member 8 |
| chr18_-_69671199 | 0.01 |
ENSRNOT00000082484
|
Smad4
|
SMAD family member 4 |
| chr1_-_100845392 | 0.01 |
ENSRNOT00000027436
|
Tbc1d17
|
TBC1 domain family, member 17 |
| chrX_-_15504165 | 0.01 |
ENSRNOT00000006233
|
Otud5
|
OTU deubiquitinase 5 |
| chr18_+_61258911 | 0.01 |
ENSRNOT00000082403
|
Zfp532
|
zinc finger protein 532 |
| chr10_-_13536899 | 0.01 |
ENSRNOT00000008537
|
Amdhd2
|
amidohydrolase domain containing 2 |
| chr7_+_80750725 | 0.01 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr2_-_198706428 | 0.01 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr10_+_98706960 | 0.01 |
ENSRNOT00000006217
|
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr6_-_95890325 | 0.01 |
ENSRNOT00000050217
|
LOC690335
|
similar to 60S ribosomal protein L23a |
| chr6_+_137243185 | 0.01 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr20_-_27308069 | 0.01 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr20_-_31598118 | 0.01 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr15_-_18675431 | 0.01 |
ENSRNOT00000080794
|
Abhd6
|
abhydrolase domain containing 6 |
| chr14_+_17228856 | 0.01 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr17_-_27969433 | 0.01 |
ENSRNOT00000073967
|
Nrn1
|
neuritin 1 |
| chr10_+_57040267 | 0.01 |
ENSRNOT00000026207
|
Arrb2
|
arrestin, beta 2 |
| chr11_-_782954 | 0.01 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr9_-_111220651 | 0.01 |
ENSRNOT00000016028
|
Gin1
|
gypsy retrotransposon integrase 1 |
| chr8_+_85417838 | 0.01 |
ENSRNOT00000012257
|
Ick
|
intestinal cell kinase |
| chr2_+_34546988 | 0.01 |
ENSRNOT00000072577
|
Adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr9_-_38196273 | 0.01 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr8_+_58120179 | 0.01 |
ENSRNOT00000049076
ENSRNOT00000090114 |
Npat
|
nuclear protein, co-activator of histone transcription |
| chrX_-_116638508 | 0.01 |
ENSRNOT00000030400
|
Lhfpl1
|
lipoma HMGIC fusion partner-like 1 |
| chr5_-_137238354 | 0.01 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr4_-_40136061 | 0.01 |
ENSRNOT00000009752
|
Bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr13_-_51201331 | 0.01 |
ENSRNOT00000005272
ENSRNOT00000078990 |
Tmem183a
|
transmembrane protein 183A |
| chr20_-_31597830 | 0.01 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr9_+_88493593 | 0.01 |
ENSRNOT00000020712
|
LOC102556337
|
mitochondrial fission factor-like |
| chr3_-_150064438 | 0.01 |
ENSRNOT00000086933
|
E2f1
|
E2F transcription factor 1 |
| chr8_+_57886168 | 0.01 |
ENSRNOT00000039336
|
Exph5
|
exophilin 5 |
| chr17_-_55346279 | 0.01 |
ENSRNOT00000025037
|
Svil
|
supervillin |
| chr1_-_246110218 | 0.01 |
ENSRNOT00000077544
|
Rfx3
|
regulatory factor X3 |
| chr10_+_54156649 | 0.01 |
ENSRNOT00000074718
|
Gas7
|
growth arrest specific 7 |
| chr1_+_164380577 | 0.01 |
ENSRNOT00000055321
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr5_-_128333805 | 0.01 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr1_+_265157379 | 0.01 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr1_-_204817080 | 0.01 |
ENSRNOT00000077956
|
Fam53b
|
family with sequence similarity 53, member B |
| chr10_+_86399827 | 0.01 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr1_+_256955652 | 0.01 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr2_-_119537837 | 0.01 |
ENSRNOT00000015200
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr11_+_61605937 | 0.01 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
| chr7_-_114848414 | 0.01 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr1_+_234252757 | 0.01 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr19_-_41960083 | 0.01 |
ENSRNOT00000000279
|
Zfp821
|
zinc finger protein 821 |
| chr3_+_160852164 | 0.01 |
ENSRNOT00000019127
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr4_-_168297373 | 0.01 |
ENSRNOT00000066575
|
Lrp6
|
LDL receptor related protein 6 |
| chr9_-_78607307 | 0.01 |
ENSRNOT00000057672
|
Abca12
|
ATP binding cassette subfamily A member 12 |
| chr4_-_129515435 | 0.01 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr2_-_116413043 | 0.01 |
ENSRNOT00000078148
|
Mynn
|
myoneurin |
| chr4_+_115416580 | 0.01 |
ENSRNOT00000018303
|
Atp6v1b1
|
ATPase H+ transporting V1 subunit B1 |
| chr1_+_153861948 | 0.01 |
ENSRNOT00000087067
|
Me3
|
malic enzyme 3 |
| chr3_-_64543100 | 0.01 |
ENSRNOT00000025803
|
Zfp385b
|
zinc finger protein 385B |
| chr11_-_67756799 | 0.01 |
ENSRNOT00000030975
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr4_+_119171245 | 0.01 |
ENSRNOT00000067651
|
Gkn2
|
gastrokine 2 |
| chr4_+_8166082 | 0.01 |
ENSRNOT00000076429
|
Srpk2
|
SRSF protein kinase 2 |
| chr1_-_262047332 | 0.01 |
ENSRNOT00000067274
|
Hpse2
|
heparanase 2 (inactive) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxa2_Foxa1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:2000426 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.0 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.0 | GO:0021997 | epidermal cell fate specification(GO:0009957) response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.0 | 0.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0070097 | alpha-catenin binding(GO:0045294) delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |