Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
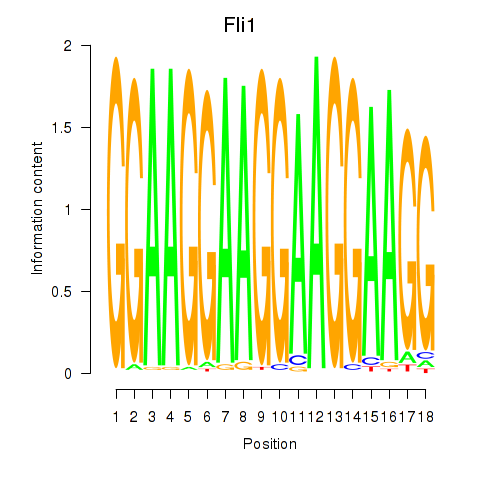
Results for Fli1
Z-value: 0.73
Transcription factors associated with Fli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fli1
|
ENSRNOG00000008904 | Fli-1 proto-oncogene, ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fli1 | rn6_v1_chr8_-_33661049_33661049 | 0.47 | 4.2e-01 | Click! |
Activity profile of Fli1 motif
Sorted Z-values of Fli1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_14292698 | 0.91 |
ENSRNOT00000073672
|
AABR07069238.1
|
|
| chr1_-_20155960 | 0.77 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr5_+_159534202 | 0.46 |
ENSRNOT00000010993
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr2_+_154604832 | 0.45 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chrX_-_123788898 | 0.44 |
ENSRNOT00000009123
|
Akap14
|
A-kinase anchoring protein 14 |
| chr1_+_150797084 | 0.41 |
ENSRNOT00000018990
|
Nox4
|
NADPH oxidase 4 |
| chr12_-_23841049 | 0.40 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr5_-_78393143 | 0.40 |
ENSRNOT00000043610
|
LOC100910017
|
60S ribosomal protein L31-like |
| chr3_+_171597241 | 0.39 |
ENSRNOT00000029558
|
LOC689618
|
similar to Protein C20orf85 homolog |
| chr9_-_17212628 | 0.38 |
ENSRNOT00000002656
ENSRNOT00000007876 |
RGD1562399
|
similar to 40S ribosomal protein S2 |
| chr13_-_109997092 | 0.37 |
ENSRNOT00000005190
|
Nenf
|
neudesin neurotrophic factor |
| chr6_+_1657331 | 0.35 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chr3_+_140106766 | 0.33 |
ENSRNOT00000014046
|
Naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr19_+_38619764 | 0.28 |
ENSRNOT00000027225
|
LOC100360573
|
ribosomal protein S12-like |
| chr4_-_153373649 | 0.28 |
ENSRNOT00000016495
|
Atp6v1e1
|
ATPase H+ transporting V1 subunit E1 |
| chr5_+_128865389 | 0.26 |
ENSRNOT00000068697
|
Calr4
|
calreticulin 4 |
| chr12_+_41636994 | 0.26 |
ENSRNOT00000001882
|
Sdsl
|
serine dehydratase-like |
| chr7_+_143754892 | 0.23 |
ENSRNOT00000085896
|
Soat2
|
sterol O-acyltransferase 2 |
| chrX_-_154653758 | 0.22 |
ENSRNOT00000090728
|
LOC103690293
|
fragile X mental retardation 1 neighbor protein-like |
| chr8_+_127144903 | 0.21 |
ENSRNOT00000013530
|
Eomes
|
eomesodermin |
| chr18_-_59819113 | 0.20 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr1_-_64021321 | 0.20 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr16_+_20020444 | 0.20 |
ENSRNOT00000024827
|
Pgls
|
6-phosphogluconolactonase |
| chr12_+_42097626 | 0.18 |
ENSRNOT00000001893
|
Tbx5
|
T-box 5 |
| chr1_-_21854763 | 0.16 |
ENSRNOT00000089196
ENSRNOT00000020528 |
Ctgf
|
connective tissue growth factor |
| chr17_+_57074525 | 0.15 |
ENSRNOT00000020012
ENSRNOT00000074146 |
Crem
|
cAMP responsive element modulator |
| chr1_-_24302298 | 0.15 |
ENSRNOT00000083452
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_-_84189266 | 0.14 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr7_-_117587103 | 0.14 |
ENSRNOT00000035423
|
Scrt1
|
scratch family transcriptional repressor 1 |
| chr14_-_72970329 | 0.14 |
ENSRNOT00000006325
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr4_+_14039977 | 0.14 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr3_+_160092975 | 0.14 |
ENSRNOT00000080707
|
Pkig
|
cAMP-dependent protein kinase inhibitor gamma |
| chr1_+_256370850 | 0.14 |
ENSRNOT00000030962
|
Cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr1_-_259287684 | 0.14 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr20_+_18492169 | 0.13 |
ENSRNOT00000046536
|
LOC103690821
|
60S ribosomal protein L39 |
| chr14_-_10694164 | 0.13 |
ENSRNOT00000080099
|
AC099384.2
|
|
| chr1_+_72874404 | 0.13 |
ENSRNOT00000058900
|
Dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr10_-_66873948 | 0.12 |
ENSRNOT00000039261
|
Evi2a
|
ecotropic viral integration site 2A |
| chr4_-_145699577 | 0.12 |
ENSRNOT00000014377
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr4_+_78735279 | 0.11 |
ENSRNOT00000011970
|
Malsu1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr7_-_2623781 | 0.11 |
ENSRNOT00000004173
|
Spryd4
|
SPRY domain containing 4 |
| chr1_-_225594958 | 0.11 |
ENSRNOT00000027492
|
Scgb1d2
|
secretoglobin, family 1D, member 2 |
| chr11_+_58624198 | 0.11 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr1_-_85317968 | 0.10 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr2_-_116372226 | 0.10 |
ENSRNOT00000011932
|
Lrriq4
|
leucine-rich repeats and IQ motif containing 4 |
| chr7_-_67345295 | 0.10 |
ENSRNOT00000087045
ENSRNOT00000005829 |
Avpr1a
|
arginine vasopressin receptor 1A |
| chr1_-_8878136 | 0.09 |
ENSRNOT00000064836
ENSRNOT00000075850 |
Vta1
|
vesicle trafficking 1 |
| chr14_+_108826831 | 0.09 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr2_+_116416507 | 0.09 |
ENSRNOT00000030700
|
Actrt3
|
actin-related protein T3 |
| chr4_+_172942020 | 0.09 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr3_-_67668772 | 0.09 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr4_+_180291389 | 0.08 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr16_+_7758996 | 0.08 |
ENSRNOT00000061063
|
Btd
|
biotinidase |
| chr7_+_23913830 | 0.08 |
ENSRNOT00000006570
|
Rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr6_-_127653124 | 0.08 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr17_+_49322205 | 0.08 |
ENSRNOT00000017713
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr2_-_144323254 | 0.08 |
ENSRNOT00000018780
|
Sertm1
|
serine-rich and transmembrane domain containing 1 |
| chr9_-_117867557 | 0.08 |
ENSRNOT00000056321
|
AABR07068760.1
|
|
| chr7_+_13881583 | 0.07 |
ENSRNOT00000065282
|
Cyp4f37
|
cytochrome P450, family 4, subfamily f, polypeptide 37 |
| chr4_-_170932618 | 0.07 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr20_-_6556350 | 0.07 |
ENSRNOT00000035819
|
Lemd2
|
LEM domain containing 2 |
| chr20_+_48881194 | 0.07 |
ENSRNOT00000000304
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr20_-_32353677 | 0.07 |
ENSRNOT00000035355
|
Stox1
|
storkhead box 1 |
| chr1_+_80383050 | 0.07 |
ENSRNOT00000023451
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr1_-_91074294 | 0.07 |
ENSRNOT00000075236
|
LOC687508
|
similar to Cytochrome c oxidase polypeptide VIIa-heart, mitochondrial precursor (Cytochrome c oxidase subunit VIIa-H) (COX VIIa-M) |
| chr3_-_176816114 | 0.07 |
ENSRNOT00000079262
ENSRNOT00000018697 |
Stmn3
|
stathmin 3 |
| chr12_+_22449985 | 0.07 |
ENSRNOT00000075951
|
Slc12a9
|
solute carrier family 12, member 9 |
| chr16_-_39996338 | 0.06 |
ENSRNOT00000089665
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chrX_+_15378789 | 0.06 |
ENSRNOT00000029272
|
Gata1
|
GATA binding protein 1 |
| chr7_+_107467260 | 0.06 |
ENSRNOT00000009240
ENSRNOT00000077382 |
Tg
|
thyroglobulin |
| chr5_-_136098013 | 0.06 |
ENSRNOT00000089166
|
RGD1563714
|
RGD1563714 |
| chr2_-_250600517 | 0.06 |
ENSRNOT00000016872
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr5_+_164751440 | 0.06 |
ENSRNOT00000010558
|
RGD1305350
|
similar to RIKEN cDNA 2510039O18 |
| chr17_+_47302272 | 0.06 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chr3_+_123731539 | 0.05 |
ENSRNOT00000051064
|
Cdc25b
|
cell division cycle 25B |
| chr1_-_52495582 | 0.05 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chrX_+_156355376 | 0.05 |
ENSRNOT00000078304
|
Lage3
|
L antigen family, member 3 |
| chr6_+_93385457 | 0.05 |
ENSRNOT00000010300
|
Actr10
|
actin-related protein 10 homolog |
| chr1_+_88586837 | 0.05 |
ENSRNOT00000080555
|
Zfp382
|
zinc finger protein 382 |
| chr5_+_133864798 | 0.05 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr8_-_62458301 | 0.05 |
ENSRNOT00000021653
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2 |
| chr2_+_250600823 | 0.05 |
ENSRNOT00000083750
|
Sep15
|
selenoprotein 15 |
| chr3_-_24601063 | 0.05 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr20_-_11528332 | 0.05 |
ENSRNOT00000038419
|
Tspear
|
thrombospondin-type laminin G domain and EAR repeats |
| chr2_-_189903219 | 0.05 |
ENSRNOT00000017000
|
S100a1
|
S100 calcium binding protein A1 |
| chr12_+_25264400 | 0.05 |
ENSRNOT00000087656
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr18_+_63098144 | 0.05 |
ENSRNOT00000024968
|
Cidea
|
cell death-inducing DFFA-like effector a |
| chr1_+_190200324 | 0.05 |
ENSRNOT00000001933
|
Abca16
|
ATP-binding cassette, subfamily A (ABC1), member 16 |
| chr2_-_183250928 | 0.04 |
ENSRNOT00000081080
|
AABR07012039.1
|
|
| chr9_+_81427730 | 0.04 |
ENSRNOT00000019109
ENSRNOT00000081711 |
Cxcr2
|
C-X-C motif chemokine receptor 2 |
| chr10_+_106991935 | 0.04 |
ENSRNOT00000004007
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr7_+_3133506 | 0.04 |
ENSRNOT00000049972
|
Pmel
|
premelanosome protein |
| chr2_+_205488182 | 0.04 |
ENSRNOT00000024270
|
Sike1
|
suppressor of IKBKE 1 |
| chr17_-_14828329 | 0.04 |
ENSRNOT00000022830
|
Barx1
|
BARX homeobox 1 |
| chr12_+_25264192 | 0.04 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chrX_+_43625169 | 0.04 |
ENSRNOT00000086311
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr10_-_61576952 | 0.04 |
ENSRNOT00000092712
|
Pafah1b1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 |
| chr18_+_52550739 | 0.04 |
ENSRNOT00000037529
|
Ctxn3
|
cortexin 3 |
| chr8_+_25246292 | 0.04 |
ENSRNOT00000021625
|
Npsr1
|
neuropeptide S receptor 1 |
| chr1_+_91433030 | 0.03 |
ENSRNOT00000015205
|
Slc7a10
|
solute carrier family 7 member 10 |
| chr3_-_7203420 | 0.03 |
ENSRNOT00000015236
|
Gfi1b
|
growth factor independent 1B transcriptional repressor |
| chr14_+_70780623 | 0.03 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr15_-_45550285 | 0.03 |
ENSRNOT00000012948
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr4_+_156050054 | 0.03 |
ENSRNOT00000039855
|
Clec4a
|
C-type lectin domain family 4, member A |
| chr7_+_137680530 | 0.03 |
ENSRNOT00000006970
|
Arid2
|
AT-rich interaction domain 2 |
| chr8_-_79862974 | 0.03 |
ENSRNOT00000091386
|
AC128582.1
|
|
| chr5_-_135628750 | 0.03 |
ENSRNOT00000079236
|
AC126292.3
|
|
| chr4_-_158705885 | 0.02 |
ENSRNOT00000087115
ENSRNOT00000026685 |
Ntf3
|
neurotrophin 3 |
| chr4_-_163528220 | 0.02 |
ENSRNOT00000090813
|
Klri1
|
killer cell lectin-like receptor family I member 1 |
| chr12_-_38805121 | 0.02 |
ENSRNOT00000057716
|
Psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr5_-_160179978 | 0.02 |
ENSRNOT00000022820
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr16_-_69884962 | 0.02 |
ENSRNOT00000078635
|
Cnga2
|
cyclic nucleotide gated channel alpha 2 |
| chr3_+_11657370 | 0.02 |
ENSRNOT00000073840
|
Ak1
|
adenylate kinase 1 |
| chr1_-_220787238 | 0.02 |
ENSRNOT00000083656
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr17_+_6684621 | 0.02 |
ENSRNOT00000089138
|
RGD1311345
|
similar to CG9752-PA |
| chr19_-_39087880 | 0.02 |
ENSRNOT00000070822
|
Chtf8
|
chromosome transmission fidelity factor 8 |
| chr18_+_55666027 | 0.02 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr1_-_80594136 | 0.02 |
ENSRNOT00000024800
|
Apoc2
|
apolipoprotein C2 |
| chr3_+_100770975 | 0.02 |
ENSRNOT00000089233
|
Bdnf
|
brain-derived neurotrophic factor |
| chr10_+_46840113 | 0.01 |
ENSRNOT00000086083
ENSRNOT00000079133 |
Myo15a
|
myosin XVA |
| chr1_-_146029840 | 0.01 |
ENSRNOT00000016455
|
Mesdc1
|
mesoderm development candidate 1 |
| chr4_-_51199570 | 0.01 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr1_+_248723397 | 0.01 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr3_+_43255567 | 0.01 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr10_-_82887497 | 0.01 |
ENSRNOT00000005644
|
Itga3
|
integrin subunit alpha 3 |
| chr15_-_20783063 | 0.01 |
ENSRNOT00000083268
|
Bmp4
|
bone morphogenetic protein 4 |
| chr11_-_71136673 | 0.01 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr9_-_24383679 | 0.01 |
ENSRNOT00000040542
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr1_+_212558257 | 0.00 |
ENSRNOT00000024912
|
Prap1
|
proline-rich acidic protein 1 |
| chr5_-_150390099 | 0.00 |
ENSRNOT00000014479
|
Ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr6_-_111572949 | 0.00 |
ENSRNOT00000016884
|
RGD1565693
|
similar to GLE1-like, RNA export mediator isoform 1 |
| chr14_+_100415668 | 0.00 |
ENSRNOT00000008057
|
C1d
|
C1D nuclear receptor co-repressor |
| chr5_+_173237642 | 0.00 |
ENSRNOT00000068770
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr5_+_137257637 | 0.00 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr13_-_72869396 | 0.00 |
ENSRNOT00000093563
|
RGD1304622
|
similar to 6820428L09 protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fli1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903544 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0045819 | cellular response to water deprivation(GO:0042631) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.4 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.0 | 0.5 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |