Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
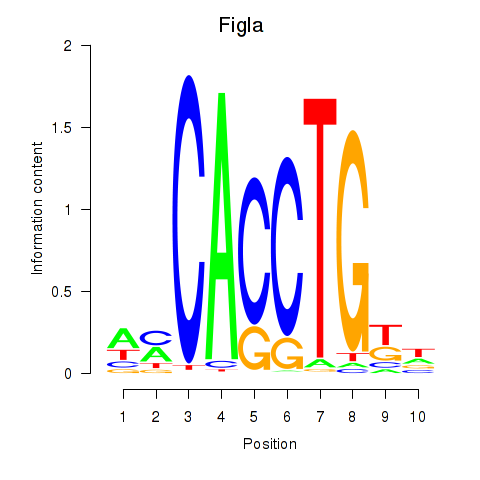
Results for Figla
Z-value: 0.28
Transcription factors associated with Figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Figla
|
ENSRNOG00000015877 | folliculogenesis specific bHLH transcription factor |
Activity profile of Figla motif
Sorted Z-values of Figla motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_27083410 | 0.16 |
ENSRNOT00000000432
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chrX_+_111396995 | 0.08 |
ENSRNOT00000087634
|
Pih1d3
|
PIH1 domain containing 3 |
| chr20_+_5050327 | 0.07 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr4_-_28437676 | 0.07 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr8_+_116754178 | 0.06 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr6_-_59950586 | 0.06 |
ENSRNOT00000005800
|
Arl4a
|
ADP-ribosylation factor like GTPase 4A |
| chr4_-_482645 | 0.06 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr1_+_80135391 | 0.06 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr1_-_221431713 | 0.05 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr10_-_14013870 | 0.05 |
ENSRNOT00000004172
|
Slc9a3r2
|
SLC9A3 regulator 2 |
| chr1_+_100891866 | 0.05 |
ENSRNOT00000086890
ENSRNOT00000083336 |
Fuz
|
fuzzy planar cell polarity protein |
| chr10_+_104368247 | 0.05 |
ENSRNOT00000006519
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr1_+_41325462 | 0.05 |
ENSRNOT00000081017
ENSRNOT00000078494 ENSRNOT00000088168 |
Esr1
|
estrogen receptor 1 |
| chr2_+_95008311 | 0.04 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chr9_+_66335492 | 0.04 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr16_-_49574314 | 0.04 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr15_+_4475051 | 0.04 |
ENSRNOT00000008461
|
Kcnk16
|
potassium two pore domain channel subfamily K member 16 |
| chr7_+_120273117 | 0.04 |
ENSRNOT00000014349
|
Galr3
|
galanin receptor 3 |
| chr16_-_39970532 | 0.04 |
ENSRNOT00000071331
|
Spata4
|
spermatogenesis associated 4 |
| chr3_+_148386189 | 0.04 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr13_+_80517536 | 0.03 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr7_-_143016040 | 0.03 |
ENSRNOT00000029697
|
Krt80
|
keratin 80 |
| chr8_-_117353672 | 0.03 |
ENSRNOT00000027262
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr14_+_33354807 | 0.03 |
ENSRNOT00000049774
|
Hopx
|
HOP homeobox |
| chr4_-_82271893 | 0.03 |
ENSRNOT00000075005
|
Hoxa7
|
homeobox A7 |
| chr6_+_26051396 | 0.03 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr5_-_155772040 | 0.03 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr10_-_82887497 | 0.03 |
ENSRNOT00000005644
|
Itga3
|
integrin subunit alpha 3 |
| chr5_+_154522119 | 0.03 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr7_-_2912491 | 0.03 |
ENSRNOT00000048453
|
AC128207.1
|
|
| chr18_+_61377051 | 0.03 |
ENSRNOT00000066659
|
Oacyl
|
O-acyltransferase like |
| chr10_+_86303727 | 0.03 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr19_+_22699808 | 0.03 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr15_+_48674380 | 0.03 |
ENSRNOT00000018762
|
Fbxo16
|
F-box protein 16 |
| chr15_+_89407426 | 0.03 |
ENSRNOT00000039423
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr8_+_32018560 | 0.03 |
ENSRNOT00000007358
|
Adamts8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr4_-_82186190 | 0.02 |
ENSRNOT00000071729
|
LOC100911622
|
homeobox protein Hox-A7-like |
| chr3_+_161236898 | 0.02 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr5_+_29538380 | 0.02 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr4_-_60358562 | 0.02 |
ENSRNOT00000018001
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr1_+_7252349 | 0.02 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chrX_-_154918095 | 0.02 |
ENSRNOT00000085224
|
LOC100911991
|
elongation factor 1-alpha 1-like |
| chr1_-_165680176 | 0.02 |
ENSRNOT00000025245
ENSRNOT00000082697 |
Plekhb1
|
pleckstrin homology domain containing B1 |
| chr10_-_89088993 | 0.02 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chr10_-_108196217 | 0.02 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr15_-_37383277 | 0.02 |
ENSRNOT00000011711
|
Gjb2
|
gap junction protein, beta 2 |
| chr3_-_60460724 | 0.02 |
ENSRNOT00000024706
|
Chrna1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr18_+_48132414 | 0.02 |
ENSRNOT00000050631
|
Snx2
|
sorting nexin 2 |
| chr1_+_85368288 | 0.02 |
ENSRNOT00000026720
|
Med29
|
mediator complex subunit 29 |
| chr13_-_110642754 | 0.02 |
ENSRNOT00000029039
|
LOC100362350
|
hydroxysteroid 17-beta dehydrogenase 6-like |
| chr5_-_24631679 | 0.02 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr10_-_87529599 | 0.02 |
ENSRNOT00000074099
|
Krtap2-1
|
keratin associated protein 2-1 |
| chr10_-_54467956 | 0.02 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr2_-_43068677 | 0.02 |
ENSRNOT00000036658
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr1_+_100470722 | 0.02 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr9_-_18371856 | 0.02 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr17_-_42267079 | 0.02 |
ENSRNOT00000059564
|
LOC100911664
|
uncharacterized LOC100911664 |
| chr10_+_106812739 | 0.02 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr20_-_2210033 | 0.02 |
ENSRNOT00000084596
|
Trim26
|
tripartite motif-containing 26 |
| chr1_+_80000165 | 0.02 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr3_+_112531703 | 0.01 |
ENSRNOT00000041727
|
LOC100911204
|
protein CASC5-like |
| chrX_+_92131209 | 0.01 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr10_+_65772443 | 0.01 |
ENSRNOT00000013296
|
Sebox
|
SEBOX homeobox |
| chr10_+_16542882 | 0.01 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr6_-_26051229 | 0.01 |
ENSRNOT00000005855
|
Bre
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr1_-_80311043 | 0.01 |
ENSRNOT00000068092
|
Klc3
|
kinesin light chain 3 |
| chr10_-_87535438 | 0.01 |
ENSRNOT00000086873
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr3_+_11476883 | 0.01 |
ENSRNOT00000072241
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr1_-_64433636 | 0.01 |
ENSRNOT00000080032
|
Prkcg
|
protein kinase C, gamma |
| chr16_-_40025401 | 0.01 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr1_+_78739930 | 0.01 |
ENSRNOT00000021976
|
Strn4
|
striatin 4 |
| chr20_+_6102277 | 0.01 |
ENSRNOT00000033064
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr8_+_62341613 | 0.01 |
ENSRNOT00000066923
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr10_-_66848388 | 0.01 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr9_+_99998275 | 0.01 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr10_-_70220558 | 0.01 |
ENSRNOT00000041389
ENSRNOT00000076398 ENSRNOT00000076596 |
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr1_-_72464492 | 0.01 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chr4_-_52350624 | 0.01 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
| chr10_-_87521514 | 0.01 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr9_+_94286550 | 0.01 |
ENSRNOT00000026504
|
Chrnd
|
cholinergic receptor nicotinic delta subunit |
| chr8_+_39997875 | 0.01 |
ENSRNOT00000050609
|
Esam
|
endothelial cell adhesion molecule |
| chr18_+_3887419 | 0.01 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr10_-_56289882 | 0.01 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr4_-_39102807 | 0.01 |
ENSRNOT00000052063
|
Thsd7a
|
thrombospondin type 1 domain containing 7A |
| chr12_-_47360507 | 0.01 |
ENSRNOT00000001562
|
Sppl3
|
signal peptide peptidase like 3 |
| chr4_+_56981283 | 0.01 |
ENSRNOT00000010989
|
Tspan33
|
tetraspanin 33 |
| chr8_-_22874637 | 0.01 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr20_-_2701637 | 0.00 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr3_+_125503638 | 0.00 |
ENSRNOT00000028900
|
Crls1
|
cardiolipin synthase 1 |
| chr6_-_11494459 | 0.00 |
ENSRNOT00000021570
|
Kcnk12
|
potassium two pore domain channel subfamily K member 12 |
| chr17_+_47721977 | 0.00 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr13_+_70157522 | 0.00 |
ENSRNOT00000036906
|
Apobec4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr8_+_55504359 | 0.00 |
ENSRNOT00000059086
|
Btg4
|
BTG anti-proliferation factor 4 |
| chr11_+_50781127 | 0.00 |
ENSRNOT00000002738
|
Alcam
|
activated leukocyte cell adhesion molecule |
| chr3_-_129357348 | 0.00 |
ENSRNOT00000084829
ENSRNOT00000007410 |
Pak7
|
p21 (RAC1) activated kinase 7 |
| chr11_-_35749464 | 0.00 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr10_+_87808493 | 0.00 |
ENSRNOT00000065405
|
LOC680428
|
hypothetical protein LOC680428 |
| chr7_-_101140308 | 0.00 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr2_-_184289126 | 0.00 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Figla
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.0 | GO:1990375 | baculum development(GO:1990375) |
| 0.0 | 0.0 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |