Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Evx2
Z-value: 0.68
Transcription factors associated with Evx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Evx2
|
ENSRNOG00000001589 | even-skipped homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Evx2 | rn6_v1_chr3_-_61581598_61581598 | -0.06 | 9.2e-01 | Click! |
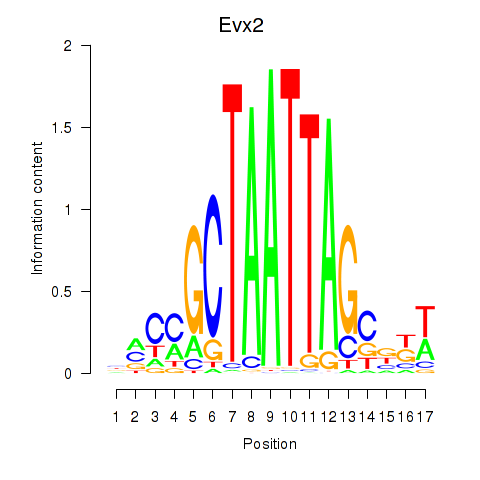
Activity profile of Evx2 motif
Sorted Z-values of Evx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88670430 | 2.23 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr12_-_2174131 | 1.28 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr4_+_148782479 | 1.04 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr20_-_9855443 | 0.46 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr7_-_28711761 | 0.40 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr9_+_98924134 | 0.29 |
ENSRNOT00000027597
|
Twist2
|
twist family bHLH transcription factor 2 |
| chr20_+_44680449 | 0.26 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr17_-_43776460 | 0.18 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr13_+_89480058 | 0.16 |
ENSRNOT00000050804
|
Cfap126
|
cilia and flagella associated protein 126 |
| chr2_+_188583664 | 0.16 |
ENSRNOT00000045477
|
Dpm3
|
dolichyl-phosphate mannosyltransferase subunit 3 |
| chr8_+_57983556 | 0.15 |
ENSRNOT00000009562
|
RGD1311251
|
similar to RIKEN cDNA 4930550C14 |
| chr14_-_115052450 | 0.15 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr19_-_37938857 | 0.14 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr12_-_19167015 | 0.14 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr10_+_103395511 | 0.14 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr3_+_80072489 | 0.11 |
ENSRNOT00000057176
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr5_+_28485619 | 0.10 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr5_-_151473750 | 0.09 |
ENSRNOT00000011357
|
Tmem222
|
transmembrane protein 222 |
| chr13_+_90514336 | 0.09 |
ENSRNOT00000088996
ENSRNOT00000085377 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr12_-_48627297 | 0.08 |
ENSRNOT00000000890
|
Iscu
|
iron-sulfur cluster assembly enzyme |
| chr1_-_189238776 | 0.08 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr16_+_51748970 | 0.08 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr13_-_73638073 | 0.08 |
ENSRNOT00000005195
|
Cep350
|
centrosomal protein 350 |
| chr7_+_38900593 | 0.08 |
ENSRNOT00000072166
|
Epyc
|
epiphycan |
| chr3_-_165537940 | 0.08 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr10_+_11046221 | 0.08 |
ENSRNOT00000005109
|
Nmral1
|
NmrA like redox sensor 1 |
| chr13_+_90943255 | 0.08 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr9_+_15166118 | 0.07 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr11_-_61234944 | 0.07 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr16_-_70242979 | 0.07 |
ENSRNOT00000071269
|
LOC103689964
|
vacuolar ATPase assembly integral membrane protein Vma21 |
| chr1_+_189432604 | 0.07 |
ENSRNOT00000034610
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr4_+_70977556 | 0.06 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr7_-_12311149 | 0.06 |
ENSRNOT00000046886
|
Dazap1
|
DAZ associated protein 1 |
| chr4_-_21920651 | 0.06 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr1_+_185863043 | 0.06 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr17_-_34945317 | 0.06 |
ENSRNOT00000090457
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chrX_-_134866210 | 0.06 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr1_-_88657845 | 0.05 |
ENSRNOT00000074211
|
LOC100912380
|
calpain small subunit 1-like |
| chr20_+_7409401 | 0.05 |
ENSRNOT00000000586
|
Snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr3_+_113319456 | 0.05 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr11_-_87403367 | 0.05 |
ENSRNOT00000057751
|
Thap7
|
THAP domain containing 7 |
| chr10_-_14324170 | 0.05 |
ENSRNOT00000035513
ENSRNOT00000090587 |
Hn1l
|
hematological and neurological expressed 1-like |
| chr17_+_9797907 | 0.05 |
ENSRNOT00000021638
|
Lman2
|
lectin, mannose-binding 2 |
| chr1_-_57518458 | 0.05 |
ENSRNOT00000002040
|
Pdcd2
|
programmed cell death 2 |
| chr1_-_252461461 | 0.04 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr15_-_12889102 | 0.04 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chr20_-_157665 | 0.04 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr13_+_82438697 | 0.03 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr8_+_126975833 | 0.03 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr11_-_62067655 | 0.03 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_113091770 | 0.03 |
ENSRNOT00000068132
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr2_-_219262901 | 0.03 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr5_+_117583502 | 0.03 |
ENSRNOT00000010933
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr20_-_4698718 | 0.03 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr2_-_158156444 | 0.03 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr11_+_58624198 | 0.03 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr8_-_114617466 | 0.02 |
ENSRNOT00000082992
ENSRNOT00000038160 |
Col6a5
|
collagen type VI alpha 5 chain |
| chrX_+_110016995 | 0.02 |
ENSRNOT00000093542
|
Nrk
|
Nik related kinase |
| chr1_-_142759882 | 0.02 |
ENSRNOT00000015218
|
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr9_+_8052210 | 0.02 |
ENSRNOT00000073659
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr1_+_98414226 | 0.02 |
ENSRNOT00000090785
|
Siglecl1
|
SIGLEC family like 1 |
| chr10_-_43931094 | 0.01 |
ENSRNOT00000039778
|
Olr1415
|
olfactory receptor 1415 |
| chr15_-_12129220 | 0.01 |
ENSRNOT00000029318
|
Olr1605
|
olfactory receptor 1605 |
| chr7_-_113228439 | 0.01 |
ENSRNOT00000034672
|
Col22a1
|
collagen type XXII alpha 1 chain |
| chr18_+_16544508 | 0.01 |
ENSRNOT00000020601
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr18_-_16543992 | 0.01 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr12_+_41486076 | 0.01 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr20_+_3176107 | 0.01 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr1_-_81550598 | 0.00 |
ENSRNOT00000051671
|
RGD1564380
|
similar to BC049730 protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Evx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 2.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |