Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
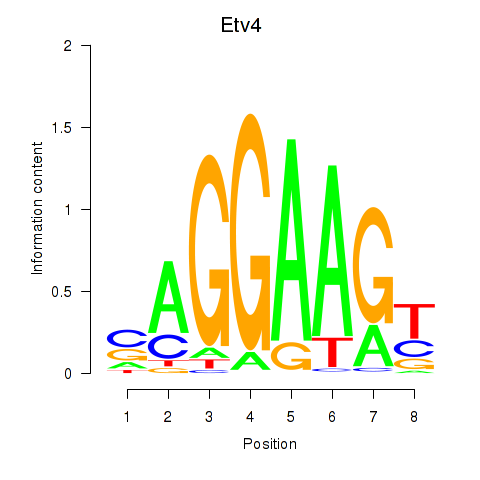
Results for Etv4
Z-value: 0.36
Transcription factors associated with Etv4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv4
|
ENSRNOG00000020792 | ets variant 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv4 | rn6_v1_chr10_-_89700283_89700283 | 0.57 | 3.2e-01 | Click! |
Activity profile of Etv4 motif
Sorted Z-values of Etv4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_190003223 | 0.27 |
ENSRNOT00000015712
|
S100a5
|
S100 calcium binding protein A5 |
| chr10_+_83655460 | 0.15 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr10_-_64642292 | 0.13 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr10_-_64657089 | 0.12 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr15_-_57805184 | 0.12 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr6_-_108796124 | 0.11 |
ENSRNOT00000086545
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr2_-_190100276 | 0.11 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr11_+_61531416 | 0.11 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr14_-_84393421 | 0.11 |
ENSRNOT00000006911
|
Ccdc157
|
coiled-coil domain containing 157 |
| chr12_-_47987255 | 0.10 |
ENSRNOT00000074000
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr10_+_85301875 | 0.10 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr10_-_83655182 | 0.10 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr7_+_127081978 | 0.09 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr3_-_43117337 | 0.09 |
ENSRNOT00000078001
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr3_+_122114108 | 0.09 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chr3_-_164239250 | 0.08 |
ENSRNOT00000012604
|
Spata2
|
spermatogenesis associated 2 |
| chr14_+_104250617 | 0.08 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr3_+_58965552 | 0.08 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr2_-_21698937 | 0.08 |
ENSRNOT00000080165
|
AABR07007642.1
|
|
| chrX_+_82143789 | 0.07 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr1_-_226887156 | 0.07 |
ENSRNOT00000054809
ENSRNOT00000028347 |
Cd6
|
Cd6 molecule |
| chr1_-_198316882 | 0.07 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr7_-_36408588 | 0.07 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr6_-_1319541 | 0.07 |
ENSRNOT00000006527
|
Strn
|
striatin |
| chr7_+_143707237 | 0.07 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr7_-_93826665 | 0.07 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr9_-_30844199 | 0.07 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr13_-_37287458 | 0.07 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr10_+_15538872 | 0.07 |
ENSRNOT00000060078
|
Luc7l
|
LUC7-like |
| chr12_+_38144855 | 0.06 |
ENSRNOT00000032274
|
Hcar1
|
hydroxycarboxylic acid receptor 1 |
| chr4_+_163162211 | 0.06 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr10_-_103685844 | 0.06 |
ENSRNOT00000064284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chrX_-_73778595 | 0.06 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr7_-_122329443 | 0.06 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr3_+_79918969 | 0.06 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr11_-_71368440 | 0.06 |
ENSRNOT00000085568
ENSRNOT00000084768 |
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr14_+_84393182 | 0.06 |
ENSRNOT00000008355
|
Sf3a1
|
splicing factor 3a, subunit 1 |
| chr1_+_8310577 | 0.06 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr2_+_41157823 | 0.06 |
ENSRNOT00000066384
|
Pde4d
|
phosphodiesterase 4D |
| chr16_+_71058022 | 0.06 |
ENSRNOT00000066901
|
Bag4
|
BCL2-associated athanogene 4 |
| chr5_+_82587420 | 0.06 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr20_+_4355175 | 0.06 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr1_+_276309927 | 0.06 |
ENSRNOT00000067460
ENSRNOT00000066236 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr13_-_88536728 | 0.06 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr1_-_221015929 | 0.06 |
ENSRNOT00000028137
|
Sipa1
|
signal-induced proliferation-associated 1 |
| chr8_-_21968415 | 0.06 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr7_+_145068286 | 0.06 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr10_-_14299167 | 0.06 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr1_+_88078350 | 0.06 |
ENSRNOT00000048677
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr4_-_63039422 | 0.06 |
ENSRNOT00000015808
|
Mtpn
|
myotrophin |
| chr4_+_27473477 | 0.06 |
ENSRNOT00000007940
ENSRNOT00000079571 |
Ankib1
|
ankyrin repeat and IBR domain containing 1 |
| chr4_-_27473150 | 0.06 |
ENSRNOT00000032505
|
Krit1
|
KRIT1, ankyrin repeat containing |
| chr2_-_243224883 | 0.06 |
ENSRNOT00000014139
|
Dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr17_-_78812111 | 0.05 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr6_-_76552559 | 0.05 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr19_-_11451278 | 0.05 |
ENSRNOT00000026118
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr1_-_257498844 | 0.05 |
ENSRNOT00000019056
|
Noc3l
|
NOC3-like DNA replication regulator |
| chr7_-_11223649 | 0.05 |
ENSRNOT00000061191
|
Mfsd12
|
major facilitator superfamily domain containing 12 |
| chr4_+_34282625 | 0.05 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chr1_-_260254600 | 0.05 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr14_-_82171480 | 0.05 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr3_-_113423473 | 0.05 |
ENSRNOT00000064982
|
Serinc4
|
serine incorporator 4 |
| chr2_-_200003443 | 0.05 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr10_+_91254058 | 0.05 |
ENSRNOT00000087218
ENSRNOT00000065373 |
Fmnl1
|
formin-like 1 |
| chr10_+_89646195 | 0.05 |
ENSRNOT00000048140
|
Dhx8
|
DEAH-box helicase 8 |
| chr12_+_48628278 | 0.05 |
ENSRNOT00000000891
|
Sart3
|
squamous cell carcinoma antigen recognized by T-cells 3 |
| chr7_+_12782491 | 0.05 |
ENSRNOT00000065093
|
Cnn2
|
calponin 2 |
| chr10_+_75087892 | 0.05 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chr6_+_86713803 | 0.05 |
ENSRNOT00000005861
|
Fam179b
|
family with sequence similarity 179, member B |
| chr10_-_13892997 | 0.05 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr3_+_80555196 | 0.05 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr9_+_73687743 | 0.05 |
ENSRNOT00000079495
ENSRNOT00000017197 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr11_+_84745904 | 0.05 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr11_-_74793673 | 0.05 |
ENSRNOT00000002338
ENSRNOT00000002343 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr14_+_8182383 | 0.05 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr18_+_29629184 | 0.05 |
ENSRNOT00000021797
|
Hars2
|
histidyl-tRNA synthetase 2 |
| chr20_-_5441706 | 0.05 |
ENSRNOT00000000549
|
Vps52
|
VPS52 GARP complex subunit |
| chr11_-_66759402 | 0.05 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr10_-_109333899 | 0.05 |
ENSRNOT00000006225
|
Slc38a10
|
solute carrier family 38, member 10 |
| chr10_+_47018974 | 0.05 |
ENSRNOT00000079375
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr6_+_134844676 | 0.05 |
ENSRNOT00000007595
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr18_-_1389929 | 0.04 |
ENSRNOT00000051362
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr10_-_88754829 | 0.04 |
ENSRNOT00000026354
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr2_+_208373154 | 0.04 |
ENSRNOT00000082943
ENSRNOT00000050538 |
LOC100911796
|
adenosine receptor A3-like |
| chr5_+_144031402 | 0.04 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr8_-_62424303 | 0.04 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr10_+_94988362 | 0.04 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr20_+_5646097 | 0.04 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr7_+_132857628 | 0.04 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr4_+_147037179 | 0.04 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr5_-_137321121 | 0.04 |
ENSRNOT00000027414
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr13_-_55173692 | 0.04 |
ENSRNOT00000064785
ENSRNOT00000029878 ENSRNOT00000029865 ENSRNOT00000060292 ENSRNOT00000000814 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr5_-_147635789 | 0.04 |
ENSRNOT00000037106
|
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr3_+_11679530 | 0.04 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr6_-_86713370 | 0.04 |
ENSRNOT00000005821
|
Klhl28
|
kelch-like family member 28 |
| chr6_+_104291071 | 0.04 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr3_+_161272385 | 0.04 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr3_+_160231914 | 0.04 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr9_+_94745217 | 0.04 |
ENSRNOT00000051338
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr18_-_1390190 | 0.04 |
ENSRNOT00000061777
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr13_+_68785827 | 0.04 |
ENSRNOT00000003517
|
Trmt1l
|
tRNA methyltransferase 1-like |
| chr8_-_133002201 | 0.04 |
ENSRNOT00000008772
|
Ccr1
|
C-C motif chemokine receptor 1 |
| chr3_+_107142762 | 0.04 |
ENSRNOT00000006177
|
RGD1563680
|
similar to CDNA sequence BC052040 |
| chr6_+_86713604 | 0.04 |
ENSRNOT00000059271
|
Fam179b
|
family with sequence similarity 179, member B |
| chr3_-_122946289 | 0.04 |
ENSRNOT00000055810
|
Pced1a
|
PC-esterase domain containing 1A |
| chr1_+_43475589 | 0.04 |
ENSRNOT00000092034
ENSRNOT00000024682 |
Oprm1
|
opioid receptor, mu 1 |
| chr10_+_34383396 | 0.04 |
ENSRNOT00000047186
|
Olr1386
|
olfactory receptor 1386 |
| chrX_+_134979646 | 0.04 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr3_-_112676556 | 0.04 |
ENSRNOT00000014664
|
Cdan1
|
codanin 1 |
| chrX_+_10964067 | 0.04 |
ENSRNOT00000093181
ENSRNOT00000066480 |
Med14
|
mediator complex subunit 14 |
| chr5_+_25042710 | 0.04 |
ENSRNOT00000061385
|
RGD1559904
|
similar to mKIAA1429 protein |
| chr11_+_64601029 | 0.03 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr3_+_67849966 | 0.03 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr13_+_104816371 | 0.03 |
ENSRNOT00000086497
|
AABR07022057.1
|
|
| chr3_-_80875817 | 0.03 |
ENSRNOT00000091265
|
Dgkz
|
diacylglycerol kinase zeta |
| chr11_-_30428073 | 0.03 |
ENSRNOT00000047741
|
Scaf4
|
SR-related CTD-associated factor 4 |
| chr13_+_99005142 | 0.03 |
ENSRNOT00000004293
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr18_+_22964210 | 0.03 |
ENSRNOT00000066816
|
Pik3c3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr15_-_34469350 | 0.03 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr1_+_219403970 | 0.03 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr9_+_82718709 | 0.03 |
ENSRNOT00000027256
ENSRNOT00000080524 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr20_+_5125349 | 0.03 |
ENSRNOT00000085598
ENSRNOT00000001129 |
Bag6
|
BCL2-associated athanogene 6 |
| chr10_-_56270640 | 0.03 |
ENSRNOT00000056918
|
Cd68
|
Cd68 molecule |
| chr7_-_144778656 | 0.03 |
ENSRNOT00000055290
|
Smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr12_-_52558606 | 0.03 |
ENSRNOT00000056656
|
Golga3
|
golgin A3 |
| chr5_+_144160108 | 0.03 |
ENSRNOT00000064972
|
Eva1b
|
eva-1 homolog B |
| chr18_+_56379890 | 0.03 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr13_-_68785760 | 0.03 |
ENSRNOT00000086460
ENSRNOT00000044255 |
Swt1
|
SWT1 RNA endoribonuclease homolog |
| chr3_-_162579201 | 0.03 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr1_-_44474674 | 0.03 |
ENSRNOT00000078768
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr12_-_6956914 | 0.03 |
ENSRNOT00000072129
ENSRNOT00000001210 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr10_-_34166599 | 0.03 |
ENSRNOT00000003246
|
Trim41
|
tripartite motif-containing 41 |
| chr18_-_24735349 | 0.03 |
ENSRNOT00000036537
|
Gpr17
|
G protein-coupled receptor 17 |
| chr10_+_12046541 | 0.03 |
ENSRNOT00000081191
|
Mefv
|
MEFV, pyrin innate immunity regulator |
| chrX_-_123515720 | 0.03 |
ENSRNOT00000092343
|
Nkrf
|
NFKB repressing factor |
| chr2_-_33025271 | 0.03 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_+_173660921 | 0.03 |
ENSRNOT00000066561
|
Noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr10_+_18996523 | 0.03 |
ENSRNOT00000046135
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr14_-_86333424 | 0.03 |
ENSRNOT00000083191
|
Nudcd3
|
NudC domain containing 3 |
| chr10_-_90995982 | 0.03 |
ENSRNOT00000093266
|
Gfap
|
glial fibrillary acidic protein |
| chr1_+_175839108 | 0.03 |
ENSRNOT00000023741
|
Ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr12_-_43940798 | 0.03 |
ENSRNOT00000001485
|
RGD1562310
|
similar to hypothetical protein FLJ21415 |
| chr2_-_139528162 | 0.03 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr13_+_37400476 | 0.03 |
ENSRNOT00000003435
|
Ccdc93
|
coiled-coil domain containing 93 |
| chr5_-_141242131 | 0.03 |
ENSRNOT00000081482
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr2_-_187133993 | 0.03 |
ENSRNOT00000019502
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr12_+_13734429 | 0.03 |
ENSRNOT00000001479
|
Fbxl18
|
F-box and leucine-rich repeat protein 18 |
| chr16_+_6970342 | 0.03 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr7_+_120206188 | 0.03 |
ENSRNOT00000086853
|
Triobp
|
TRIO and F-actin binding protein |
| chr7_+_130227456 | 0.03 |
ENSRNOT00000078423
ENSRNOT00000011070 |
Ppp6r2
|
protein phosphatase 6, regulatory subunit 2 |
| chr6_-_26241337 | 0.03 |
ENSRNOT00000006525
|
Slc4a1ap
|
solute carrier family 4 member 1 adaptor protein |
| chr10_+_12046701 | 0.03 |
ENSRNOT00000011073
ENSRNOT00000084004 |
Mefv
|
MEFV, pyrin innate immunity regulator |
| chr3_+_113423693 | 0.03 |
ENSRNOT00000021091
|
Hypk
|
Huntingtin interacting protein K |
| chr13_+_47602692 | 0.03 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr5_+_126254142 | 0.02 |
ENSRNOT00000057471
|
Pars2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr2_+_202470487 | 0.02 |
ENSRNOT00000026953
|
Gdap2
|
ganglioside-induced differentiation-associated-protein 2 |
| chr1_+_81779380 | 0.02 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chr5_-_137238354 | 0.02 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr2_-_120316600 | 0.02 |
ENSRNOT00000084521
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr2_-_227582895 | 0.02 |
ENSRNOT00000020574
|
Mettl14
|
methyltransferase like 14 |
| chr3_-_11102515 | 0.02 |
ENSRNOT00000035580
|
Fam78a
|
family with sequence similarity 78, member A |
| chr20_+_3300247 | 0.02 |
ENSRNOT00000035838
|
Prr3
|
proline rich 3 |
| chr2_-_206997519 | 0.02 |
ENSRNOT00000027042
|
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr20_-_5455632 | 0.02 |
ENSRNOT00000000552
|
Wdr46
|
WD repeat domain 46 |
| chr1_+_170242846 | 0.02 |
ENSRNOT00000023751
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr9_+_94879745 | 0.02 |
ENSRNOT00000080482
|
Atg16l1
|
autophagy related 16-like 1 |
| chr13_-_100740728 | 0.02 |
ENSRNOT00000000074
|
Fbxo28
|
F-box protein 28 |
| chr6_-_132958546 | 0.02 |
ENSRNOT00000041903
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr8_+_4440876 | 0.02 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr2_-_18927365 | 0.02 |
ENSRNOT00000045850
|
Xrcc4
|
X-ray repair cross complementing 4 |
| chr2_+_181331464 | 0.02 |
ENSRNOT00000017448
|
Map9
|
microtubule-associated protein 9 |
| chr3_+_104899682 | 0.02 |
ENSRNOT00000041910
|
Fmn1
|
formin 1 |
| chr7_+_121889157 | 0.02 |
ENSRNOT00000024835
|
Fam83f
|
family with sequence similarity 83, member F |
| chr5_-_159602251 | 0.02 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chr4_-_119327822 | 0.02 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr2_-_227582594 | 0.02 |
ENSRNOT00000065222
|
Mettl14
|
methyltransferase like 14 |
| chr7_+_127964752 | 0.02 |
ENSRNOT00000038168
|
RGD1560617
|
hypothetical gene supported by NM_053561; AF062594 |
| chr5_+_64294321 | 0.02 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr10_-_88645364 | 0.02 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr11_-_60882379 | 0.02 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr2_-_120316357 | 0.02 |
ENSRNOT00000065469
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr3_+_97721668 | 0.02 |
ENSRNOT00000065168
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr13_+_48790767 | 0.02 |
ENSRNOT00000087504
|
Elk4
|
ELK4, ETS transcription factor |
| chr8_+_115179893 | 0.02 |
ENSRNOT00000017469
|
Rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr2_-_184993341 | 0.02 |
ENSRNOT00000071580
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr9_-_81565416 | 0.02 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr3_-_162581610 | 0.02 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr10_+_61746082 | 0.02 |
ENSRNOT00000003992
|
Tsr1
|
TSR1, ribosome maturation factor |
| chr5_+_142702685 | 0.02 |
ENSRNOT00000085986
ENSRNOT00000010373 ENSRNOT00000087416 |
Sf3a3
|
splicing factor 3a, subunit 3 |
| chr5_-_142681173 | 0.02 |
ENSRNOT00000009882
|
Utp11
|
UTP11, small subunit processome component homolog (S. cerevisiae) |
| chr10_-_12916784 | 0.02 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr8_+_76400341 | 0.02 |
ENSRNOT00000087382
ENSRNOT00000081072 |
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr4_+_21862313 | 0.02 |
ENSRNOT00000007948
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr1_+_215628785 | 0.02 |
ENSRNOT00000054864
|
Lsp1
|
lymphocyte-specific protein 1 |
| chr1_-_126211439 | 0.02 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr1_+_20022793 | 0.02 |
ENSRNOT00000061404
|
L3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_-_110517163 | 0.02 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr6_-_50786967 | 0.02 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr2_-_53300404 | 0.02 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr1_-_198662610 | 0.02 |
ENSRNOT00000055012
|
Sept1
|
septin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:2000426 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0070428 | detection of lipopolysaccharide(GO:0032497) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.0 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.0 | 0.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.0 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.0 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.0 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0043533 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0051425 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |