Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
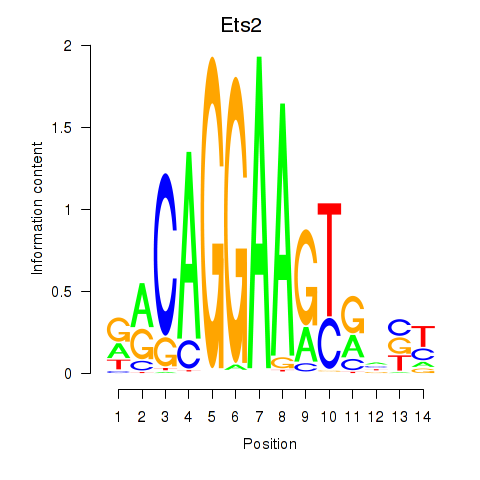
Results for Ets2
Z-value: 0.37
Transcription factors associated with Ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ets2
|
ENSRNOG00000001647 | ETS proto-oncogene 2, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ets2 | rn6_v1_chr11_+_36075709_36075709 | -0.07 | 9.1e-01 | Click! |
Activity profile of Ets2 motif
Sorted Z-values of Ets2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_36408588 | 0.18 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_-_93826665 | 0.16 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr12_+_38144855 | 0.13 |
ENSRNOT00000032274
|
Hcar1
|
hydroxycarboxylic acid receptor 1 |
| chr1_+_276309927 | 0.12 |
ENSRNOT00000067460
ENSRNOT00000066236 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr8_+_45797315 | 0.12 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr11_+_84745904 | 0.11 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr13_+_89774764 | 0.11 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr13_-_100740728 | 0.10 |
ENSRNOT00000000074
|
Fbxo28
|
F-box protein 28 |
| chr3_-_111037425 | 0.10 |
ENSRNOT00000085628
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr16_-_49820235 | 0.09 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr10_+_72197977 | 0.08 |
ENSRNOT00000003886
|
Myo19
|
myosin XIX |
| chr1_-_198120061 | 0.08 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr5_+_58661049 | 0.08 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr2_+_118746333 | 0.08 |
ENSRNOT00000079431
|
AABR07009965.1
|
|
| chr3_+_58965552 | 0.08 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr17_+_13670520 | 0.07 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr9_+_81880177 | 0.07 |
ENSRNOT00000022839
|
Stk36
|
serine/threonine kinase 36 |
| chr7_+_58366192 | 0.07 |
ENSRNOT00000081891
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr5_-_147761983 | 0.07 |
ENSRNOT00000012936
|
Lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr18_-_56115593 | 0.07 |
ENSRNOT00000045041
|
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr5_-_79222687 | 0.07 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr3_-_2474913 | 0.07 |
ENSRNOT00000014445
|
Ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr4_+_157523770 | 0.06 |
ENSRNOT00000055985
ENSRNOT00000023240 |
Zfp384
|
zinc finger protein 384 |
| chr4_-_155116154 | 0.06 |
ENSRNOT00000020529
|
Phc1
|
polyhomeotic homolog 1 |
| chr10_+_94988362 | 0.06 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr1_+_266844480 | 0.06 |
ENSRNOT00000027482
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr15_+_61069581 | 0.06 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr1_+_224882439 | 0.06 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr1_-_169456098 | 0.06 |
ENSRNOT00000030827
|
Trim30c
|
tripartite motif-containing 30C |
| chr14_+_70780623 | 0.06 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr15_-_57805184 | 0.06 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr5_-_169630340 | 0.06 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr13_-_78957183 | 0.06 |
ENSRNOT00000076954
ENSRNOT00000003851 |
Klhl20
|
kelch-like family member 20 |
| chr10_-_64642292 | 0.06 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr8_+_33239139 | 0.06 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr4_+_157523320 | 0.06 |
ENSRNOT00000023192
|
Zfp384
|
zinc finger protein 384 |
| chr13_-_52514875 | 0.06 |
ENSRNOT00000064758
|
Nav1
|
neuron navigator 1 |
| chr9_-_116222374 | 0.06 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr8_+_107882219 | 0.05 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr3_-_59688692 | 0.05 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr20_-_5485837 | 0.05 |
ENSRNOT00000092272
ENSRNOT00000000559 ENSRNOT00000092597 |
Daxx
|
death-domain associated protein |
| chr6_-_8956276 | 0.05 |
ENSRNOT00000079027
|
Six2
|
SIX homeobox 2 |
| chr14_-_84393421 | 0.05 |
ENSRNOT00000006911
|
Ccdc157
|
coiled-coil domain containing 157 |
| chr14_+_84393182 | 0.05 |
ENSRNOT00000008355
|
Sf3a1
|
splicing factor 3a, subunit 1 |
| chr1_+_81779380 | 0.05 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chrX_-_123515720 | 0.05 |
ENSRNOT00000092343
|
Nkrf
|
NFKB repressing factor |
| chr7_-_142180997 | 0.05 |
ENSRNOT00000087632
|
Tfcp2
|
transcription factor CP2 |
| chr7_-_2712723 | 0.05 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr5_+_60250546 | 0.05 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr6_+_107169528 | 0.05 |
ENSRNOT00000012495
|
Psen1
|
presenilin 1 |
| chr6_+_86651196 | 0.05 |
ENSRNOT00000034756
|
RGD1307621
|
hypothetical LOC314168 |
| chr17_-_78812111 | 0.05 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr13_-_67206688 | 0.05 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr1_-_234670113 | 0.05 |
ENSRNOT00000017133
|
LOC499331
|
similar to hypothetical protein D030056L22 |
| chr2_-_188745144 | 0.05 |
ENSRNOT00000055533
|
Cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr4_+_157524423 | 0.04 |
ENSRNOT00000036654
|
Zfp384
|
zinc finger protein 384 |
| chr8_+_55196758 | 0.04 |
ENSRNOT00000065879
|
Fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr1_-_92119951 | 0.04 |
ENSRNOT00000018153
ENSRNOT00000092121 |
Zfp507
|
zinc finger protein 507 |
| chr1_+_87938042 | 0.04 |
ENSRNOT00000027837
|
Map4k1
|
mitogen activated protein kinase kinase kinase kinase 1 |
| chr1_-_175420106 | 0.04 |
ENSRNOT00000013126
ENSRNOT00000077125 |
Sbf2
|
SET binding factor 2 |
| chr4_-_56897310 | 0.04 |
ENSRNOT00000043902
ENSRNOT00000090038 |
Tnpo3
|
transportin 3 |
| chr10_-_92602082 | 0.04 |
ENSRNOT00000007963
|
Cdc27
|
cell division cycle 27 |
| chr15_-_42898150 | 0.04 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr2_+_188745503 | 0.04 |
ENSRNOT00000056652
|
Shc1
|
SHC adaptor protein 1 |
| chr1_+_104106245 | 0.04 |
ENSRNOT00000019175
ENSRNOT00000081660 |
Zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr5_-_113532878 | 0.04 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr3_-_72602548 | 0.04 |
ENSRNOT00000031745
|
Lrrc55
|
leucine rich repeat containing 55 |
| chrX_+_68752597 | 0.04 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr9_-_30844199 | 0.04 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr5_-_48194251 | 0.04 |
ENSRNOT00000081748
ENSRNOT00000065912 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr10_+_15538872 | 0.04 |
ENSRNOT00000060078
|
Luc7l
|
LUC7-like |
| chr18_-_786674 | 0.04 |
ENSRNOT00000021955
|
Cetn1
|
centrin 1 |
| chr3_+_93909156 | 0.04 |
ENSRNOT00000090365
|
Lmo2
|
LIM domain only 2 |
| chr7_+_2795901 | 0.04 |
ENSRNOT00000047462
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr9_+_82393672 | 0.04 |
ENSRNOT00000032419
|
Ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr10_-_34166599 | 0.04 |
ENSRNOT00000003246
|
Trim41
|
tripartite motif-containing 41 |
| chr8_+_77107536 | 0.04 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chr8_-_13835168 | 0.04 |
ENSRNOT00000014435
|
Med17
|
mediator complex subunit 17 |
| chr11_-_1819094 | 0.04 |
ENSRNOT00000000911
|
MGC95208
|
similar to 4930453N24Rik protein |
| chrX_+_144807392 | 0.04 |
ENSRNOT00000004529
|
NEWGENE_1565644
|
leucine zipper down-regulated in cancer 1 |
| chr5_+_82587420 | 0.04 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr2_+_181331464 | 0.04 |
ENSRNOT00000017448
|
Map9
|
microtubule-associated protein 9 |
| chr10_-_31419235 | 0.04 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_-_22281778 | 0.04 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr7_-_14302552 | 0.04 |
ENSRNOT00000091368
|
Brd4
|
bromodomain containing 4 |
| chr7_-_142180794 | 0.04 |
ENSRNOT00000037447
|
Tfcp2
|
transcription factor CP2 |
| chr1_-_281101438 | 0.04 |
ENSRNOT00000012734
|
Rab11fip2
|
RAB11 family interacting protein 2 |
| chr5_-_163119208 | 0.04 |
ENSRNOT00000085291
ENSRNOT00000022185 |
Vps13d
|
vacuolar protein sorting 13D |
| chr1_+_13595295 | 0.04 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr3_+_80555196 | 0.04 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr2_+_202470487 | 0.04 |
ENSRNOT00000026953
|
Gdap2
|
ganglioside-induced differentiation-associated-protein 2 |
| chr4_+_78320190 | 0.04 |
ENSRNOT00000032742
ENSRNOT00000091359 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr2_+_58534476 | 0.04 |
ENSRNOT00000077646
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr1_+_228337767 | 0.04 |
ENSRNOT00000066247
|
Patl1
|
PAT1 homolog 1, processing body mRNA decay factor |
| chr1_-_43638161 | 0.04 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr16_-_21362955 | 0.04 |
ENSRNOT00000039607
|
Gmip
|
Gem-interacting protein |
| chr4_+_176565694 | 0.04 |
ENSRNOT00000016794
|
Pyroxd1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr17_+_9746485 | 0.04 |
ENSRNOT00000072298
|
Pfn3
|
profilin 3 |
| chr10_-_70802782 | 0.04 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr5_-_137238354 | 0.03 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr3_+_55960067 | 0.03 |
ENSRNOT00000010216
|
Ppig
|
peptidylprolyl isomerase G |
| chr16_-_71057883 | 0.03 |
ENSRNOT00000020721
|
Lsm1
|
LSM1 homolog, mRNA degradation associated |
| chr1_+_167169442 | 0.03 |
ENSRNOT00000048172
|
LOC102549471
|
short transient receptor potential channel 2-like |
| chr17_-_10001901 | 0.03 |
ENSRNOT00000082836
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr1_-_89509343 | 0.03 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr5_-_166430254 | 0.03 |
ENSRNOT00000048914
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr3_+_55960327 | 0.03 |
ENSRNOT00000086584
|
Ppig
|
peptidylprolyl isomerase G |
| chrX_+_82143789 | 0.03 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr3_-_164239250 | 0.03 |
ENSRNOT00000012604
|
Spata2
|
spermatogenesis associated 2 |
| chr14_+_78939903 | 0.03 |
ENSRNOT00000078562
ENSRNOT00000088221 |
Man2b2
Mrfap1
|
mannosidase, alpha, class 2B, member 2 Morf4 family associated protein 1 |
| chr15_-_4399589 | 0.03 |
ENSRNOT00000065858
ENSRNOT00000079812 |
Fam149b1
|
family with sequence similarity 149, member B1 |
| chr16_+_74177215 | 0.03 |
ENSRNOT00000025851
|
Ikbkb
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr2_+_115678344 | 0.03 |
ENSRNOT00000036717
|
Slc7a14
|
solute carrier family 7, member 14 |
| chr2_+_41157823 | 0.03 |
ENSRNOT00000066384
|
Pde4d
|
phosphodiesterase 4D |
| chr2_+_4942775 | 0.03 |
ENSRNOT00000093548
ENSRNOT00000093741 |
Fam172a
|
family with sequence similarity 172, member A |
| chr4_+_152892388 | 0.03 |
ENSRNOT00000056198
ENSRNOT00000090218 ENSRNOT00000075895 |
Kdm5a
|
lysine demethylase 5A |
| chr2_-_118882562 | 0.03 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr8_+_70522092 | 0.03 |
ENSRNOT00000025873
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr10_-_85084850 | 0.03 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr12_+_12374790 | 0.03 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr1_+_80256973 | 0.03 |
ENSRNOT00000024113
|
Ercc1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr5_-_146787676 | 0.03 |
ENSRNOT00000008887
|
Zscan20
|
zinc finger and SCAN domain containing 20 |
| chr16_-_47874856 | 0.03 |
ENSRNOT00000077810
|
Rwdd4
|
RWD domain containing 4 |
| chr14_+_1463359 | 0.03 |
ENSRNOT00000070834
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr2_+_234375315 | 0.03 |
ENSRNOT00000071270
|
LOC102549542
|
elongation of very long chain fatty acids protein 6-like |
| chr15_+_52451161 | 0.03 |
ENSRNOT00000018725
|
Dok2
|
docking protein 2 |
| chr8_-_49271834 | 0.03 |
ENSRNOT00000085022
|
Ube4a
|
ubiquitination factor E4A |
| chr8_-_21968415 | 0.03 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr16_+_4469468 | 0.03 |
ENSRNOT00000021164
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr5_-_135472116 | 0.03 |
ENSRNOT00000022170
|
Nasp
|
nuclear autoantigenic sperm protein |
| chr2_-_120316357 | 0.03 |
ENSRNOT00000065469
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr14_-_86796378 | 0.03 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr8_+_109455786 | 0.03 |
ENSRNOT00000039593
|
Msl2
|
male-specific lethal 2 homolog (Drosophila) |
| chr4_+_113101100 | 0.03 |
ENSRNOT00000084897
|
AABR07061261.1
|
|
| chr15_-_33333417 | 0.03 |
ENSRNOT00000018982
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr1_-_199064150 | 0.03 |
ENSRNOT00000044511
|
Zfp689
|
zinc finger protein 689 |
| chr5_+_150459713 | 0.03 |
ENSRNOT00000081681
ENSRNOT00000074251 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chr8_-_22625959 | 0.03 |
ENSRNOT00000012138
|
Yipf2
|
Yip1 domain family, member 2 |
| chr20_+_28815039 | 0.03 |
ENSRNOT00000075800
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr1_+_189550354 | 0.03 |
ENSRNOT00000083153
|
Exnef
|
exonuclease NEF-sp |
| chr13_-_47979797 | 0.03 |
ENSRNOT00000080035
|
Rassf5
|
Ras association domain family member 5 |
| chr9_-_113598477 | 0.03 |
ENSRNOT00000035606
ENSRNOT00000084884 |
Ralbp1
|
ralA binding protein 1 |
| chr1_-_189549891 | 0.03 |
ENSRNOT00000019747
|
Eri2
|
ERI1 exoribonuclease family member 2 |
| chr9_-_10801140 | 0.03 |
ENSRNOT00000072771
|
Fem1a
|
fem-1 homolog A |
| chr7_+_3246220 | 0.03 |
ENSRNOT00000009155
|
Sarnp
|
SAP domain containing ribonucleoprotein |
| chr19_-_52252587 | 0.03 |
ENSRNOT00000020990
|
Taf1c
|
TATA-box binding protein associated factor, RNA polymerase 1 subunit C |
| chr19_+_45938915 | 0.03 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr1_-_123064642 | 0.03 |
ENSRNOT00000013551
|
Mkrn3
|
makorin, ring finger protein, 3 |
| chr2_-_62634785 | 0.03 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr7_+_28414350 | 0.03 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr10_-_13892997 | 0.03 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr4_-_55527092 | 0.02 |
ENSRNOT00000087407
ENSRNOT00000010223 |
Zfp800
|
zinc finger protein 800 |
| chr2_-_20919824 | 0.02 |
ENSRNOT00000086300
|
Zcchc9
|
zinc finger CCHC-type containing 9 |
| chr2_+_237148941 | 0.02 |
ENSRNOT00000015116
|
Dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr20_-_45399694 | 0.02 |
ENSRNOT00000000715
|
Amd1
|
adenosylmethionine decarboxylase 1 |
| chr13_+_99136871 | 0.02 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr18_+_57654290 | 0.02 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr3_-_111037166 | 0.02 |
ENSRNOT00000017070
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr8_+_64088913 | 0.02 |
ENSRNOT00000013022
|
Adpgk
|
ADP-dependent glucokinase |
| chr13_-_85443727 | 0.02 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr11_+_61531416 | 0.02 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr3_+_152909189 | 0.02 |
ENSRNOT00000066341
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr13_-_88642011 | 0.02 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr2_+_187993758 | 0.02 |
ENSRNOT00000027182
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chrX_-_157474219 | 0.02 |
ENSRNOT00000079860
|
Zfp275
|
zinc finger protein 275 |
| chr3_+_60024013 | 0.02 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr1_+_114258719 | 0.02 |
ENSRNOT00000088459
ENSRNOT00000016376 |
Cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr1_+_174655939 | 0.02 |
ENSRNOT00000014002
|
Ipo7
|
importin 7 |
| chr4_+_157523110 | 0.02 |
ENSRNOT00000081640
|
Zfp384
|
zinc finger protein 384 |
| chr4_+_123760743 | 0.02 |
ENSRNOT00000013498
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr1_-_199624783 | 0.02 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr10_-_109802739 | 0.02 |
ENSRNOT00000054951
|
Sirt7
|
sirtuin 7 |
| chr5_+_122847645 | 0.02 |
ENSRNOT00000009516
|
Oma1
|
OMA1 zinc metallopeptidase |
| chr1_+_23977688 | 0.02 |
ENSRNOT00000014805
|
Tbpl1
|
TATA-box binding protein like 1 |
| chr8_-_123371257 | 0.02 |
ENSRNOT00000017243
|
Stt3b
|
STT3B, catalytic subunit of the oligosaccharyltransferase complex |
| chr1_-_260254600 | 0.02 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr9_+_14551758 | 0.02 |
ENSRNOT00000017157
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr7_-_14303055 | 0.02 |
ENSRNOT00000008963
|
Brd4
|
bromodomain containing 4 |
| chr2_+_57206613 | 0.02 |
ENSRNOT00000082694
ENSRNOT00000046069 |
Nup155
|
nucleoporin 155 |
| chr2_+_263864331 | 0.02 |
ENSRNOT00000055259
|
Zranb2
|
zinc finger RANBP2-type containing 2 |
| chr1_-_256013495 | 0.02 |
ENSRNOT00000023342
|
Ide
|
insulin degrading enzyme |
| chr3_+_28627084 | 0.02 |
ENSRNOT00000049884
|
Arhgap15
|
Rho GTPase activating protein 15 |
| chr12_+_19231092 | 0.02 |
ENSRNOT00000045379
|
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr1_+_197098603 | 0.02 |
ENSRNOT00000022365
|
LOC361646
|
similar to K04F10.2 |
| chr18_-_37096132 | 0.02 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr13_+_80125391 | 0.02 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr3_-_110517163 | 0.02 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr15_+_57221292 | 0.02 |
ENSRNOT00000014502
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr4_+_9882904 | 0.02 |
ENSRNOT00000016909
|
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr8_-_71533069 | 0.02 |
ENSRNOT00000021863
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr4_+_152892599 | 0.02 |
ENSRNOT00000079090
|
Kdm5a
|
lysine demethylase 5A |
| chr17_+_66446569 | 0.02 |
ENSRNOT00000070825
|
Heatr1
|
HEAT repeat containing 1 |
| chr16_+_10727571 | 0.02 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr6_-_36940868 | 0.02 |
ENSRNOT00000006470
|
Gen1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr2_-_152824547 | 0.02 |
ENSRNOT00000019824
|
Dhx36
|
DEAH-box helicase 36 |
| chr2_-_243224883 | 0.02 |
ENSRNOT00000014139
|
Dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr6_-_3355339 | 0.02 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr11_+_24263281 | 0.02 |
ENSRNOT00000086946
|
Gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr9_+_65614142 | 0.02 |
ENSRNOT00000016613
|
Casp8
|
caspase 8 |
| chr9_+_94880053 | 0.02 |
ENSRNOT00000024445
|
Atg16l1
|
autophagy related 16-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ets2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 0.0 | 0.0 | GO:0051142 | positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.0 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |