Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
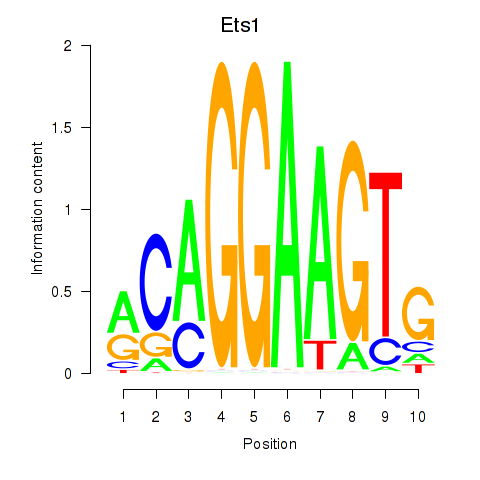
Results for Ets1
Z-value: 0.79
Transcription factors associated with Ets1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ets1
|
ENSRNOG00000008941 | ETS proto-oncogene 1, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ets1 | rn6_v1_chr8_+_33816386_33816386 | 0.62 | 2.6e-01 | Click! |
Activity profile of Ets1 motif
Sorted Z-values of Ets1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_73778595 | 0.49 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr10_-_64642292 | 0.45 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr6_+_104291071 | 0.45 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr8_-_21968415 | 0.44 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr2_+_190003223 | 0.38 |
ENSRNOT00000015712
|
S100a5
|
S100 calcium binding protein A5 |
| chr3_-_152382529 | 0.38 |
ENSRNOT00000027076
|
Scand1
|
SCAN domain-containing 1 |
| chr15_-_57805184 | 0.36 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr3_-_38277440 | 0.36 |
ENSRNOT00000037857
|
Stam2
|
signal transducing adaptor molecule 2 |
| chr4_-_63039422 | 0.34 |
ENSRNOT00000015808
|
Mtpn
|
myotrophin |
| chr5_-_169630340 | 0.33 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr7_+_143707237 | 0.29 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr3_+_101010899 | 0.28 |
ENSRNOT00000073258
|
Lin7c
|
lin-7 homolog C, crumbs cell polarity complex component |
| chr9_-_41337498 | 0.27 |
ENSRNOT00000039480
|
Fam168b
|
family with sequence similarity 168, member B |
| chr3_-_148932878 | 0.27 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr11_+_61531416 | 0.27 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr10_+_85301875 | 0.26 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr10_-_64657089 | 0.26 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr11_+_71151132 | 0.26 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr3_-_92242318 | 0.25 |
ENSRNOT00000007018
|
Trim44
|
tripartite motif-containing 44 |
| chr1_-_78521999 | 0.25 |
ENSRNOT00000021223
|
Arhgap35
|
Rho GTPase activating protein 35 |
| chr16_+_71058022 | 0.25 |
ENSRNOT00000066901
|
Bag4
|
BCL2-associated athanogene 4 |
| chr11_-_30428073 | 0.24 |
ENSRNOT00000047741
|
Scaf4
|
SR-related CTD-associated factor 4 |
| chr9_+_14551758 | 0.24 |
ENSRNOT00000017157
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr11_+_16826399 | 0.24 |
ENSRNOT00000050701
|
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr2_-_183582553 | 0.24 |
ENSRNOT00000014236
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr1_-_72339395 | 0.24 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr13_-_37287458 | 0.23 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr8_+_70522092 | 0.23 |
ENSRNOT00000025873
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr11_+_64601029 | 0.23 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr13_+_97838361 | 0.23 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr19_-_11451278 | 0.23 |
ENSRNOT00000026118
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr20_+_5646097 | 0.23 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr1_+_274245184 | 0.23 |
ENSRNOT00000018889
|
Dusp5
|
dual specificity phosphatase 5 |
| chr4_+_148139528 | 0.21 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr5_-_146973932 | 0.21 |
ENSRNOT00000007761
|
Zfp362
|
zinc finger protein 362 |
| chr5_-_173653905 | 0.20 |
ENSRNOT00000038556
|
Plekhn1
|
pleckstrin homology domain containing N1 |
| chr10_-_56530842 | 0.20 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chr17_+_15749978 | 0.20 |
ENSRNOT00000067311
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr10_-_14299167 | 0.20 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr5_-_79222687 | 0.20 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chrX_-_20070537 | 0.20 |
ENSRNOT00000093602
ENSRNOT00000003397 |
Gnl3l
|
G protein nucleolar 3 like |
| chr14_-_13058172 | 0.20 |
ENSRNOT00000002746
ENSRNOT00000071706 |
Prdm8
|
PR/SET domain 8 |
| chr15_+_37790141 | 0.20 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr3_+_71114100 | 0.19 |
ENSRNOT00000088549
ENSRNOT00000006961 |
Itgav
|
integrin subunit alpha V |
| chr10_-_85435016 | 0.19 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr1_-_198316882 | 0.19 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr6_-_86713370 | 0.19 |
ENSRNOT00000005821
|
Klhl28
|
kelch-like family member 28 |
| chr5_-_60559533 | 0.19 |
ENSRNOT00000092899
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr10_+_47018974 | 0.19 |
ENSRNOT00000079375
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr8_-_63034226 | 0.19 |
ENSRNOT00000043434
|
Pml
|
promyelocytic leukemia |
| chr14_-_86333424 | 0.18 |
ENSRNOT00000083191
|
Nudcd3
|
NudC domain containing 3 |
| chr3_+_80555196 | 0.18 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr14_-_84393421 | 0.18 |
ENSRNOT00000006911
|
Ccdc157
|
coiled-coil domain containing 157 |
| chr7_+_2795901 | 0.18 |
ENSRNOT00000047462
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr12_+_38144855 | 0.18 |
ENSRNOT00000032274
|
Hcar1
|
hydroxycarboxylic acid receptor 1 |
| chr13_-_88536728 | 0.18 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr10_+_47019326 | 0.18 |
ENSRNOT00000006984
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr20_-_5441706 | 0.18 |
ENSRNOT00000000549
|
Vps52
|
VPS52 GARP complex subunit |
| chr1_-_72311856 | 0.18 |
ENSRNOT00000021286
|
Epn1
|
Epsin 1 |
| chr3_+_11679530 | 0.17 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr4_-_155563249 | 0.17 |
ENSRNOT00000011298
|
Slc2a3
|
solute carrier family 2 member 3 |
| chr4_-_117296082 | 0.17 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr7_+_133400485 | 0.17 |
ENSRNOT00000006219
|
Cntn1
|
contactin 1 |
| chr2_-_252691886 | 0.17 |
ENSRNOT00000068739
|
Prkacb
|
protein kinase cAMP-activated catalytic subunit beta |
| chr10_-_14443010 | 0.17 |
ENSRNOT00000022142
|
Tmem204
|
transmembrane protein 204 |
| chr6_+_107169528 | 0.17 |
ENSRNOT00000012495
|
Psen1
|
presenilin 1 |
| chr19_+_45938915 | 0.17 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr6_+_28235695 | 0.17 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr4_+_163162211 | 0.17 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr4_+_157523110 | 0.17 |
ENSRNOT00000081640
|
Zfp384
|
zinc finger protein 384 |
| chr1_+_85437780 | 0.16 |
ENSRNOT00000093740
|
Supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr7_-_140245723 | 0.16 |
ENSRNOT00000088999
|
Ccnt1
|
cyclin T1 |
| chr3_-_162579201 | 0.16 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr7_-_11223649 | 0.16 |
ENSRNOT00000061191
|
Mfsd12
|
major facilitator superfamily domain containing 12 |
| chr10_+_89646195 | 0.16 |
ENSRNOT00000048140
|
Dhx8
|
DEAH-box helicase 8 |
| chr2_-_118745766 | 0.16 |
ENSRNOT00000013858
|
Zmat3
|
zinc finger, matrin type 3 |
| chr6_+_137824213 | 0.16 |
ENSRNOT00000056880
|
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr10_-_109333899 | 0.15 |
ENSRNOT00000006225
|
Slc38a10
|
solute carrier family 38, member 10 |
| chr2_-_204341676 | 0.15 |
ENSRNOT00000021713
|
Slc22a15
|
solute carrier family 22, member 15 |
| chr20_-_19637958 | 0.15 |
ENSRNOT00000074657
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chr6_-_64170122 | 0.15 |
ENSRNOT00000093248
ENSRNOT00000005363 |
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr10_+_59529785 | 0.15 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr1_+_81779380 | 0.15 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chrX_+_10964067 | 0.15 |
ENSRNOT00000093181
ENSRNOT00000066480 |
Med14
|
mediator complex subunit 14 |
| chr13_+_50196042 | 0.15 |
ENSRNOT00000090858
|
Zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr8_-_45215974 | 0.15 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr5_-_60559329 | 0.15 |
ENSRNOT00000017046
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr13_-_82607379 | 0.15 |
ENSRNOT00000051763
|
Blzf1
|
basic leucine zipper nuclear factor 1 |
| chr11_+_24263281 | 0.14 |
ENSRNOT00000086946
|
Gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr14_+_16491573 | 0.14 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr3_+_122114108 | 0.14 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chr5_+_82587420 | 0.14 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr1_-_215846911 | 0.14 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr16_-_2367253 | 0.14 |
ENSRNOT00000083573
|
Pde12
|
phosphodiesterase 12 |
| chr7_-_130151414 | 0.14 |
ENSRNOT00000010467
|
Plxnb2
|
plexin B2 |
| chr7_-_93826665 | 0.14 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr10_+_71278650 | 0.14 |
ENSRNOT00000092020
|
Synrg
|
synergin, gamma |
| chr6_+_110093819 | 0.14 |
ENSRNOT00000013602
|
Gpatch2l
|
G patch domain containing 2-like |
| chr4_+_34282625 | 0.14 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chrX_-_69218526 | 0.14 |
ENSRNOT00000092321
ENSRNOT00000074071 ENSRNOT00000092571 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr16_+_10727571 | 0.14 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr7_-_2712723 | 0.14 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr20_+_4355175 | 0.14 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr2_+_41157823 | 0.14 |
ENSRNOT00000066384
|
Pde4d
|
phosphodiesterase 4D |
| chr3_-_10269693 | 0.14 |
ENSRNOT00000021871
|
Fubp3
|
far upstream element binding protein 3 |
| chr1_-_198577226 | 0.14 |
ENSRNOT00000055013
|
Spn
|
sialophorin |
| chr3_-_43117337 | 0.14 |
ENSRNOT00000078001
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr10_-_85084850 | 0.14 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr15_-_4022223 | 0.14 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
| chr4_+_87026530 | 0.13 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr16_+_19068783 | 0.13 |
ENSRNOT00000017424
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr12_+_18074033 | 0.13 |
ENSRNOT00000001727
|
LOC103689975
|
integrator complex subunit 1-like |
| chr5_-_155316495 | 0.13 |
ENSRNOT00000017559
|
Epha8
|
Eph receptor A8 |
| chr4_-_78342863 | 0.13 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr7_+_14037620 | 0.13 |
ENSRNOT00000009606
|
Syde1
|
synapse defective Rho GTPase homolog 1 |
| chr14_+_84393182 | 0.13 |
ENSRNOT00000008355
|
Sf3a1
|
splicing factor 3a, subunit 1 |
| chr7_+_127081978 | 0.13 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr1_-_190965115 | 0.13 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr8_+_117280705 | 0.13 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr12_-_6956914 | 0.13 |
ENSRNOT00000072129
ENSRNOT00000001210 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr19_-_19832812 | 0.13 |
ENSRNOT00000084657
|
Papd5
|
poly(A) RNA polymerase D5, non-canonical |
| chr17_+_5225835 | 0.13 |
ENSRNOT00000022373
|
Zcchc6
|
zinc finger CCHC-type containing 6 |
| chr17_+_13670520 | 0.13 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr18_-_56115593 | 0.13 |
ENSRNOT00000045041
|
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr7_-_107616038 | 0.13 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr15_+_24078280 | 0.13 |
ENSRNOT00000015511
ENSRNOT00000063807 |
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr8_+_117246376 | 0.12 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr13_-_51076852 | 0.12 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr10_-_56491715 | 0.12 |
ENSRNOT00000020970
|
Kctd11
|
potassium channel tetramerization domain containing 11 |
| chr18_+_56379890 | 0.12 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr13_-_83202864 | 0.12 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr14_+_1463359 | 0.12 |
ENSRNOT00000070834
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr1_+_199037544 | 0.12 |
ENSRNOT00000025499
|
Rnf40
|
ring finger protein 40 |
| chr1_+_13595295 | 0.12 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr17_-_46794845 | 0.12 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr15_+_25933483 | 0.12 |
ENSRNOT00000018972
|
Ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr4_+_2711385 | 0.12 |
ENSRNOT00000035996
|
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr12_+_12374790 | 0.12 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr6_-_132802206 | 0.12 |
ENSRNOT00000080163
ENSRNOT00000050350 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr19_-_52252587 | 0.12 |
ENSRNOT00000020990
|
Taf1c
|
TATA-box binding protein associated factor, RNA polymerase 1 subunit C |
| chr2_-_190100276 | 0.12 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr3_+_176865156 | 0.12 |
ENSRNOT00000019084
|
Zgpat
|
zinc finger CCCH-type and G-patch domain containing |
| chr2_-_211715311 | 0.12 |
ENSRNOT00000027738
|
Prpf38b
|
pre-mRNA processing factor 38B |
| chr3_+_6211789 | 0.12 |
ENSRNOT00000012892
|
Rxra
|
retinoid X receptor alpha |
| chr3_+_95707386 | 0.12 |
ENSRNOT00000005882
|
Pax6
|
paired box 6 |
| chr19_-_54652381 | 0.12 |
ENSRNOT00000065472
|
Klhdc4
|
kelch domain containing 4 |
| chr1_+_220428481 | 0.12 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr20_+_27954433 | 0.12 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr19_-_22281778 | 0.12 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr8_+_109455786 | 0.12 |
ENSRNOT00000039593
|
Msl2
|
male-specific lethal 2 homolog (Drosophila) |
| chr10_-_13892997 | 0.11 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr10_+_43067299 | 0.11 |
ENSRNOT00000003447
|
Galnt10
|
polypeptide N-acetylgalactosaminyltransferase 10 |
| chr3_-_164239250 | 0.11 |
ENSRNOT00000012604
|
Spata2
|
spermatogenesis associated 2 |
| chr13_-_47979797 | 0.11 |
ENSRNOT00000080035
|
Rassf5
|
Ras association domain family member 5 |
| chr17_-_27112820 | 0.11 |
ENSRNOT00000018359
|
Bmp6
|
bone morphogenetic protein 6 |
| chr3_+_48096954 | 0.11 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr10_-_12916784 | 0.11 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr2_-_211831551 | 0.11 |
ENSRNOT00000039225
ENSRNOT00000090561 |
Fam102b
|
family with sequence similarity 102, member B |
| chr8_-_105462141 | 0.11 |
ENSRNOT00000066731
ENSRNOT00000078760 |
Clstn2
|
calsyntenin 2 |
| chr7_-_36408588 | 0.11 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr1_+_199197138 | 0.11 |
ENSRNOT00000065187
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chrX_+_82143789 | 0.11 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr7_-_142180997 | 0.11 |
ENSRNOT00000087632
|
Tfcp2
|
transcription factor CP2 |
| chr1_+_222746023 | 0.11 |
ENSRNOT00000028787
|
Atl3
|
atlastin GTPase 3 |
| chr1_+_81365138 | 0.11 |
ENSRNOT00000026793
|
Cadm4
|
cell adhesion molecule 4 |
| chr1_+_224882439 | 0.11 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr7_-_122403667 | 0.11 |
ENSRNOT00000088814
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr10_+_90230441 | 0.11 |
ENSRNOT00000082722
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr7_-_139907640 | 0.11 |
ENSRNOT00000045473
|
Zfp641
|
zinc finger protein 641 |
| chr6_+_86713803 | 0.11 |
ENSRNOT00000005861
|
Fam179b
|
family with sequence similarity 179, member B |
| chr10_-_90995982 | 0.11 |
ENSRNOT00000093266
|
Gfap
|
glial fibrillary acidic protein |
| chr5_-_160742479 | 0.11 |
ENSRNOT00000019447
|
Kazn
|
kazrin, periplakin interacting protein |
| chr7_-_12673659 | 0.10 |
ENSRNOT00000091650
ENSRNOT00000041277 ENSRNOT00000044865 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr15_-_56970365 | 0.10 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr8_+_4440876 | 0.10 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr18_-_31071371 | 0.10 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chrX_-_123515720 | 0.10 |
ENSRNOT00000092343
|
Nkrf
|
NFKB repressing factor |
| chr14_+_85230648 | 0.10 |
ENSRNOT00000089866
|
Ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr5_-_159602251 | 0.10 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chr6_-_79306443 | 0.10 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr8_+_49354115 | 0.10 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr8_-_128754514 | 0.10 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr5_-_113532878 | 0.10 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr18_-_76984203 | 0.10 |
ENSRNOT00000089122
|
Ctdp1
|
CTD phosphatase subunit 1 |
| chr2_-_140334912 | 0.10 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr4_+_140247313 | 0.10 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr5_+_173660921 | 0.10 |
ENSRNOT00000066561
|
Noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr20_-_6864387 | 0.10 |
ENSRNOT00000068527
|
Ppil1
|
peptidylprolyl isomerase like 1 |
| chr7_+_12022285 | 0.10 |
ENSRNOT00000024080
|
Rexo1
|
RNA exonuclease 1 homolog |
| chr3_+_119484677 | 0.10 |
ENSRNOT00000037390
|
Ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr16_-_19918644 | 0.10 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr4_-_125929002 | 0.10 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_+_157524423 | 0.10 |
ENSRNOT00000036654
|
Zfp384
|
zinc finger protein 384 |
| chr2_+_116032455 | 0.10 |
ENSRNOT00000066495
|
Phc3
|
polyhomeotic homolog 3 |
| chr3_-_80875817 | 0.09 |
ENSRNOT00000091265
|
Dgkz
|
diacylglycerol kinase zeta |
| chr10_-_15211325 | 0.09 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr8_+_62368998 | 0.09 |
ENSRNOT00000025885
|
Ulk3
|
unc-51 like kinase 3 |
| chr6_-_50786967 | 0.09 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr4_+_56625561 | 0.09 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr2_+_188139730 | 0.09 |
ENSRNOT00000056789
ENSRNOT00000077698 |
Gon4l
|
gon-4 like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ets1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.2 | 0.5 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.4 | GO:0090309 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.2 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.2 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.2 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.2 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.3 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.2 | GO:0050713 | PML body organization(GO:0030578) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0051142 | positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.2 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:0002884 | type IV hypersensitivity(GO:0001806) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0038091 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:1904844 | regulation of renal output by angiotensin(GO:0002019) response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0060687 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.2 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0071899 | odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.1 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.4 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) epidermal stem cell homeostasis(GO:0036334) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0060717 | chorion development(GO:0060717) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.0 | GO:2000836 | positive regulation of androgen secretion(GO:2000836) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.0 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0033222 | xylose binding(GO:0033222) |
| 0.1 | 0.2 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.0 | GO:0001007 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |