Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
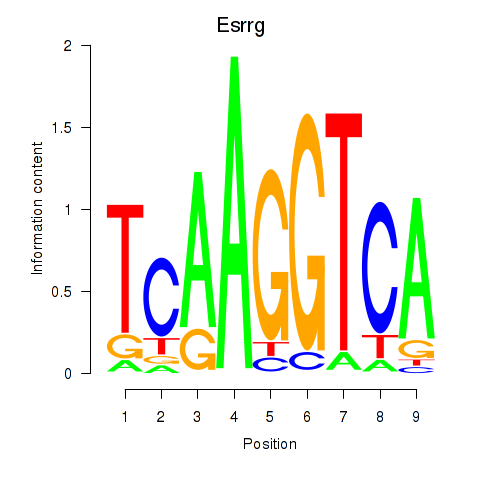
Results for Esrrg
Z-value: 0.29
Transcription factors associated with Esrrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrg
|
ENSRNOG00000002593 | estrogen-related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrg | rn6_v1_chr13_+_106463368_106463368 | 0.70 | 1.9e-01 | Click! |
Activity profile of Esrrg motif
Sorted Z-values of Esrrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 0.27 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr10_-_71849293 | 0.25 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chrX_-_40086870 | 0.17 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr3_+_147609095 | 0.11 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr4_+_51614676 | 0.10 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr8_+_59900651 | 0.10 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr4_-_130659697 | 0.10 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr12_-_30566032 | 0.07 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr5_-_75319765 | 0.07 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr7_+_129860327 | 0.06 |
ENSRNOT00000043461
|
Pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr5_-_56576676 | 0.06 |
ENSRNOT00000029712
|
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr8_+_116771982 | 0.05 |
ENSRNOT00000083240
|
AC128059.3
|
|
| chr9_-_92291220 | 0.05 |
ENSRNOT00000093357
|
Dner
|
delta/notch-like EGF repeat containing |
| chr8_+_12160067 | 0.05 |
ENSRNOT00000036338
|
Maml2
|
mastermind-like transcriptional coactivator 2 |
| chr8_-_130429132 | 0.05 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr1_+_261158261 | 0.05 |
ENSRNOT00000071965
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr20_-_5618254 | 0.04 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr5_+_50381244 | 0.04 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr8_-_68312909 | 0.03 |
ENSRNOT00000066106
|
RGD1309779
|
similar to ENSANGP00000021391 |
| chr3_+_113318563 | 0.03 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr1_+_226252226 | 0.03 |
ENSRNOT00000039369
|
AABR07006258.1
|
|
| chr6_-_107678156 | 0.03 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr8_+_106317124 | 0.03 |
ENSRNOT00000018411
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr12_+_47024442 | 0.03 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr3_+_151126591 | 0.03 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr3_-_60813869 | 0.03 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr2_+_212257225 | 0.03 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr3_+_113319456 | 0.03 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr2_+_38230757 | 0.03 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr2_-_84678790 | 0.03 |
ENSRNOT00000015886
|
Cct5
|
chaperonin containing TCP1 subunit 5 |
| chr4_-_157679962 | 0.02 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr16_+_1979191 | 0.02 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr13_-_93677377 | 0.02 |
ENSRNOT00000004917
|
Fh
|
fumarate hydratase |
| chr1_+_145770135 | 0.02 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr18_+_74156553 | 0.02 |
ENSRNOT00000022892
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr12_-_24365324 | 0.02 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr2_+_186980793 | 0.02 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr4_+_115046693 | 0.02 |
ENSRNOT00000031583
|
Bola3
|
bolA family member 3 |
| chr7_+_117409576 | 0.02 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr7_-_81592206 | 0.02 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr1_-_170404056 | 0.02 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr9_+_20213776 | 0.02 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr18_+_73564247 | 0.02 |
ENSRNOT00000077679
ENSRNOT00000035317 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr18_+_402295 | 0.02 |
ENSRNOT00000033618
|
Fundc2
|
FUN14 domain containing 2 |
| chr9_+_43259709 | 0.02 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr16_-_69242028 | 0.02 |
ENSRNOT00000019002
|
Zfp703
|
zinc finger protein 703 |
| chr9_+_90880614 | 0.02 |
ENSRNOT00000077859
ENSRNOT00000020705 ENSRNOT00000089142 |
Mff
|
mitochondrial fission factor |
| chr2_+_186980992 | 0.02 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr5_-_126911520 | 0.01 |
ENSRNOT00000091521
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr1_-_78968329 | 0.01 |
ENSRNOT00000023078
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr10_-_15155412 | 0.01 |
ENSRNOT00000026536
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr14_-_93042182 | 0.01 |
ENSRNOT00000005432
|
LOC100361025
|
protein arginine methyltransferase 1-like |
| chr12_+_25264192 | 0.01 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr11_-_60613718 | 0.01 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr13_-_85622314 | 0.01 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr10_-_88356538 | 0.01 |
ENSRNOT00000022430
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr1_-_263269762 | 0.01 |
ENSRNOT00000022309
|
Got1
|
glutamic-oxaloacetic transaminase 1 |
| chr10_+_10761477 | 0.01 |
ENSRNOT00000004176
|
Rogdi
|
rogdi homolog |
| chr6_-_123405246 | 0.01 |
ENSRNOT00000083665
|
Foxn3
|
forkhead box N3 |
| chr6_+_128750795 | 0.01 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr10_-_88357025 | 0.01 |
ENSRNOT00000080006
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr15_+_2631529 | 0.01 |
ENSRNOT00000018897
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr4_-_157331905 | 0.01 |
ENSRNOT00000020647
|
Tpi1
|
triosephosphate isomerase 1 |
| chr10_+_17486579 | 0.01 |
ENSRNOT00000080404
|
Stk10
|
serine/threonine kinase 10 |
| chr5_+_159484370 | 0.01 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr7_+_11521998 | 0.01 |
ENSRNOT00000026960
|
Sgta
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr6_+_2216623 | 0.01 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr15_+_61662264 | 0.01 |
ENSRNOT00000072969
|
Mtrf1
|
mitochondrial translation release factor 1 |
| chr3_+_94549180 | 0.01 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr16_-_29936307 | 0.01 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr5_+_156850206 | 0.01 |
ENSRNOT00000021833
|
Mul1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr18_-_27749235 | 0.01 |
ENSRNOT00000026696
|
Hspa9
|
heat shock protein family A member 9 |
| chr5_+_118743632 | 0.01 |
ENSRNOT00000013785
|
Pgm1
|
phosphoglucomutase 1 |
| chr9_+_61692154 | 0.01 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr7_-_120518653 | 0.01 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr18_+_28370085 | 0.01 |
ENSRNOT00000083993
|
Matr3
|
matrin 3 |
| chr3_+_11607225 | 0.01 |
ENSRNOT00000072685
|
St6galnac4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr10_+_64556064 | 0.01 |
ENSRNOT00000010778
|
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr1_+_190555177 | 0.01 |
ENSRNOT00000021514
|
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr10_-_82399484 | 0.00 |
ENSRNOT00000082972
ENSRNOT00000055582 |
Xylt2
|
xylosyltransferase 2 |
| chr8_+_55037750 | 0.00 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr8_+_59164572 | 0.00 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr7_+_2504695 | 0.00 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr8_-_39093277 | 0.00 |
ENSRNOT00000043344
|
LOC100911068
|
roundabout homolog 4-like |
| chr6_-_93562314 | 0.00 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr8_+_39878955 | 0.00 |
ENSRNOT00000060300
|
LOC100911068
|
roundabout homolog 4-like |
| chr11_-_71419223 | 0.00 |
ENSRNOT00000002407
|
Tfrc
|
transferrin receptor |
| chr10_-_90127600 | 0.00 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr10_+_109533487 | 0.00 |
ENSRNOT00000054975
|
Fscn2
|
fascin actin-bundling protein 2, retinal |
| chr10_+_88356615 | 0.00 |
ENSRNOT00000022687
|
Klhl10
|
kelch-like family member 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.1 | 0.3 | GO:0060066 | metanephric part of ureteric bud development(GO:0035502) oviduct development(GO:0060066) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |