Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
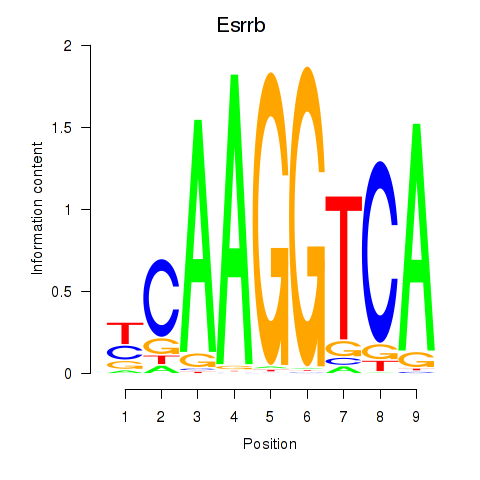
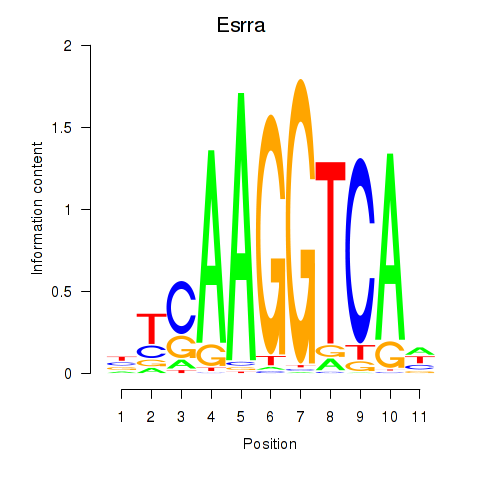
Results for Esrrb_Esrra
Z-value: 1.85
Transcription factors associated with Esrrb_Esrra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrb
|
ENSRNOG00000010259 | estrogen-related receptor beta |
|
Esrra
|
ENSRNOG00000021139 | estrogen related receptor, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrra | rn6_v1_chr1_-_222178725_222178725 | 0.51 | 3.8e-01 | Click! |
| Esrrb | rn6_v1_chr6_+_110410141_110410141 | 0.28 | 6.4e-01 | Click! |
Activity profile of Esrrb_Esrra motif
Sorted Z-values of Esrrb_Esrra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 5.49 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr10_-_71849293 | 1.56 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr20_+_14095914 | 1.37 |
ENSRNOT00000093404
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr5_+_50381244 | 1.22 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_+_276659542 | 1.18 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr10_+_110139783 | 1.15 |
ENSRNOT00000054939
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr2_-_123396147 | 1.01 |
ENSRNOT00000079004
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr2_-_14701903 | 0.96 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr19_-_37427989 | 0.95 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chrX_-_40086870 | 0.94 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr4_-_123494742 | 0.94 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr4_+_78263866 | 0.93 |
ENSRNOT00000033807
|
AI854703
|
expressed sequence AI854703 |
| chr3_+_147609095 | 0.90 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr19_-_57333433 | 0.90 |
ENSRNOT00000024917
|
Agt
|
angiotensinogen |
| chr18_-_26211445 | 0.84 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr14_+_84237520 | 0.78 |
ENSRNOT00000083580
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr6_-_6842758 | 0.77 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr12_+_47024442 | 0.76 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr10_-_15235740 | 0.74 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr1_-_167347662 | 0.68 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr1_-_167005839 | 0.68 |
ENSRNOT00000027261
|
Lrrc51
|
leucine rich repeat containing 51 |
| chr1_+_274245184 | 0.67 |
ENSRNOT00000018889
|
Dusp5
|
dual specificity phosphatase 5 |
| chr1_-_222468896 | 0.65 |
ENSRNOT00000028754
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr4_-_148845267 | 0.64 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr8_+_48569328 | 0.60 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr8_+_59900651 | 0.60 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr6_+_3012804 | 0.60 |
ENSRNOT00000061980
|
Arhgef33
|
Rho guanine nucleotide exchange factor 33 |
| chr2_-_123396386 | 0.59 |
ENSRNOT00000046700
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr4_-_165629996 | 0.59 |
ENSRNOT00000068185
ENSRNOT00000007427 |
Ybx3
|
Y box binding protein 3 |
| chr13_-_85622314 | 0.58 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr19_+_52521809 | 0.57 |
ENSRNOT00000081019
|
Klhl36
|
kelch-like family member 36 |
| chr1_-_222250980 | 0.57 |
ENSRNOT00000028734
|
Nudt22
|
nudix hydrolase 22 |
| chr9_+_43259709 | 0.57 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr4_-_123118186 | 0.56 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr12_-_24365324 | 0.56 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr5_-_56576676 | 0.56 |
ENSRNOT00000029712
|
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr8_-_130491998 | 0.55 |
ENSRNOT00000064114
|
Higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr9_+_80118029 | 0.55 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr7_+_11724962 | 0.54 |
ENSRNOT00000026551
|
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_-_60813869 | 0.54 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr14_+_43694183 | 0.53 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr1_-_167347490 | 0.53 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr2_-_119140110 | 0.52 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr7_+_35125424 | 0.52 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr3_+_2480232 | 0.52 |
ENSRNOT00000014489
|
Tprn
|
taperin |
| chr8_+_116094851 | 0.52 |
ENSRNOT00000084120
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr5_-_157573183 | 0.51 |
ENSRNOT00000064418
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr8_+_48437918 | 0.51 |
ENSRNOT00000085578
|
Mfrp
|
membrane frizzled-related protein |
| chr19_+_10596960 | 0.50 |
ENSRNOT00000021769
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr14_-_84937725 | 0.48 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr12_-_30566032 | 0.48 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr11_+_64472072 | 0.47 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr17_+_15845931 | 0.47 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr6_-_135939534 | 0.47 |
ENSRNOT00000052237
|
Diras3
|
DIRAS family GTPase 3 |
| chr20_-_3818045 | 0.47 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr12_-_44174583 | 0.46 |
ENSRNOT00000001490
|
Tesc
|
tescalcin |
| chr6_-_135412312 | 0.46 |
ENSRNOT00000010610
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr2_-_140464607 | 0.45 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr19_+_26084903 | 0.45 |
ENSRNOT00000004799
|
Prdx2
|
peroxiredoxin 2 |
| chr20_-_5618254 | 0.45 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr4_+_66405166 | 0.44 |
ENSRNOT00000077486
ENSRNOT00000067097 |
Clec2l
|
C-type lectin domain family 2, member L |
| chr15_+_41069507 | 0.43 |
ENSRNOT00000018533
|
C1qtnf9
|
C1q and tumor necrosis factor related protein 9 |
| chr19_-_54245855 | 0.43 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chrX_+_33443186 | 0.43 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr6_-_136145837 | 0.43 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr10_-_38782419 | 0.42 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr7_+_2504695 | 0.42 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr11_-_32088002 | 0.42 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr4_+_67378188 | 0.42 |
ENSRNOT00000030892
|
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr15_+_2631529 | 0.41 |
ENSRNOT00000018897
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr9_+_61692154 | 0.41 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr10_-_5533695 | 0.41 |
ENSRNOT00000051564
|
Rpl39l
|
ribosomal protein L39-like |
| chr4_-_130659697 | 0.41 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr10_-_40375605 | 0.41 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr3_+_113318563 | 0.39 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr11_-_72109964 | 0.39 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr8_-_23014499 | 0.38 |
ENSRNOT00000017820
|
Ccdc151
|
coiled-coil domain containing 151 |
| chr9_-_92291220 | 0.38 |
ENSRNOT00000093357
|
Dner
|
delta/notch-like EGF repeat containing |
| chr18_+_74156553 | 0.38 |
ENSRNOT00000022892
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr1_+_72882806 | 0.36 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr4_+_165732643 | 0.36 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr1_+_31264755 | 0.36 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr2_+_119139717 | 0.36 |
ENSRNOT00000016051
|
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr14_+_60764409 | 0.36 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr20_-_13657943 | 0.35 |
ENSRNOT00000032290
|
Vpreb3
|
pre-B lymphocyte 3 |
| chr14_-_93042182 | 0.35 |
ENSRNOT00000005432
|
LOC100361025
|
protein arginine methyltransferase 1-like |
| chr10_-_102289837 | 0.35 |
ENSRNOT00000044922
|
AABR07030729.1
|
|
| chr13_-_36101411 | 0.35 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr16_-_81243757 | 0.34 |
ENSRNOT00000024677
|
Gas6
|
growth arrest specific 6 |
| chr11_+_70687500 | 0.34 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr14_+_86673775 | 0.34 |
ENSRNOT00000091873
ENSRNOT00000079015 |
Ppia
|
peptidylprolyl isomerase A |
| chr18_+_70733872 | 0.34 |
ENSRNOT00000067018
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr10_-_91448477 | 0.34 |
ENSRNOT00000038836
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr20_-_7208292 | 0.34 |
ENSRNOT00000083169
|
Nudt3
|
nudix hydrolase 3 |
| chr11_+_37798370 | 0.34 |
ENSRNOT00000002679
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr7_+_117409576 | 0.33 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr6_+_24163026 | 0.33 |
ENSRNOT00000061284
|
Lbh
|
limb bud and heart development |
| chr19_+_54245950 | 0.32 |
ENSRNOT00000024033
|
Cox4i1
|
cytochrome c oxidase subunit 4i1 |
| chr8_+_55037750 | 0.32 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr8_+_106317124 | 0.32 |
ENSRNOT00000018411
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr6_+_128750795 | 0.32 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr1_-_253186695 | 0.32 |
ENSRNOT00000080928
|
AC096809.1
|
|
| chr14_-_79464770 | 0.32 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr2_-_260115577 | 0.32 |
ENSRNOT00000065997
|
Rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr1_+_261158261 | 0.31 |
ENSRNOT00000071965
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr1_+_221773254 | 0.31 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr8_+_117679278 | 0.31 |
ENSRNOT00000042114
|
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein I |
| chr2_-_88113029 | 0.31 |
ENSRNOT00000013354
|
Car2
|
carbonic anhydrase 2 |
| chr12_+_25264192 | 0.30 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr10_-_13107771 | 0.30 |
ENSRNOT00000005879
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr14_-_80680738 | 0.29 |
ENSRNOT00000077706
|
LOC103690128
|
smoothelin-like |
| chr5_+_150032999 | 0.29 |
ENSRNOT00000013301
|
Srsf4
|
serine and arginine rich splicing factor 4 |
| chr5_+_141560192 | 0.29 |
ENSRNOT00000023354
|
Mycbp
|
Myc binding protein |
| chr5_-_75319765 | 0.28 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr3_+_113319456 | 0.28 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr11_-_81379640 | 0.28 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_-_11018160 | 0.28 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr10_-_90356242 | 0.28 |
ENSRNOT00000028496
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr4_+_115046693 | 0.28 |
ENSRNOT00000031583
|
Bola3
|
bolA family member 3 |
| chr7_-_74735650 | 0.28 |
ENSRNOT00000014407
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chrX_-_113584459 | 0.28 |
ENSRNOT00000025923
|
Kcne5
|
potassium voltage-gated channel subfamily E regulatory subunit 5 |
| chr1_-_89488223 | 0.28 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr4_-_157331905 | 0.28 |
ENSRNOT00000020647
|
Tpi1
|
triosephosphate isomerase 1 |
| chr2_+_202200797 | 0.27 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr6_+_8669722 | 0.27 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr1_+_226252226 | 0.27 |
ENSRNOT00000039369
|
AABR07006258.1
|
|
| chr15_+_33606124 | 0.27 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr9_-_43127887 | 0.26 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr14_+_89314176 | 0.26 |
ENSRNOT00000006765
|
Upp1
|
uridine phosphorylase 1 |
| chr8_-_68312909 | 0.26 |
ENSRNOT00000066106
|
RGD1309779
|
similar to ENSANGP00000021391 |
| chr9_+_90880614 | 0.26 |
ENSRNOT00000077859
ENSRNOT00000020705 ENSRNOT00000089142 |
Mff
|
mitochondrial fission factor |
| chr12_-_40227297 | 0.26 |
ENSRNOT00000030600
|
Fam109a
|
family with sequence similarity 109, member A |
| chr9_+_20213776 | 0.26 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr7_+_129860327 | 0.26 |
ENSRNOT00000043461
|
Pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr6_+_55085313 | 0.25 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr6_+_126434226 | 0.25 |
ENSRNOT00000090857
|
Chga
|
chromogranin A |
| chr3_-_123702732 | 0.25 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr9_+_81566074 | 0.24 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr4_+_122781095 | 0.24 |
ENSRNOT00000008962
|
Hdac11
|
histone deacetylase 11 |
| chr8_+_118378059 | 0.24 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr4_-_157679962 | 0.24 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr3_-_122813583 | 0.24 |
ENSRNOT00000009681
|
Idh3B
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr9_-_16647458 | 0.24 |
ENSRNOT00000024380
|
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr20_+_18493538 | 0.24 |
ENSRNOT00000000749
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr4_-_28953067 | 0.24 |
ENSRNOT00000013989
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr16_+_19874034 | 0.23 |
ENSRNOT00000023579
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chr17_-_2705123 | 0.23 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr20_+_21316826 | 0.23 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr8_-_130429132 | 0.23 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr20_-_6961162 | 0.23 |
ENSRNOT00000000635
|
Mtch1
|
mitochondrial carrier 1 |
| chr3_+_72226613 | 0.23 |
ENSRNOT00000010619
|
Timm10
|
translocase of inner mitochondrial membrane 10 |
| chr14_-_44375804 | 0.23 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr5_+_173640780 | 0.23 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr8_-_128711221 | 0.22 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr8_-_128266639 | 0.22 |
ENSRNOT00000064555
ENSRNOT00000066932 |
Scn5a
|
sodium voltage-gated channel alpha subunit 5 |
| chr3_+_4083864 | 0.22 |
ENSRNOT00000006186
|
Fam69b
|
family with sequence similarity 69, member B |
| chr1_+_100593680 | 0.22 |
ENSRNOT00000078153
ENSRNOT00000027063 |
Kcnc3
|
potassium voltage-gated channel subfamily C member 3 |
| chr8_+_116857684 | 0.22 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr5_+_159484370 | 0.22 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr5_+_147185474 | 0.22 |
ENSRNOT00000000134
|
Ak2
|
adenylate kinase 2 |
| chr4_+_155313671 | 0.21 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chr13_-_93677377 | 0.21 |
ENSRNOT00000004917
|
Fh
|
fumarate hydratase |
| chr9_-_71830730 | 0.21 |
ENSRNOT00000019963
|
Cryga
|
crystallin, gamma A |
| chr1_+_89162639 | 0.21 |
ENSRNOT00000028508
|
Atp4a
|
ATPase H+/K+ transporting alpha subunit |
| chr15_+_4351292 | 0.20 |
ENSRNOT00000009207
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr10_+_64556064 | 0.20 |
ENSRNOT00000010778
|
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr3_-_1584946 | 0.20 |
ENSRNOT00000031058
|
Pax8
|
paired box 8 |
| chr5_+_151741817 | 0.20 |
ENSRNOT00000081318
|
Kdf1
|
keratinocyte differentiation factor 1 |
| chr13_-_95943761 | 0.20 |
ENSRNOT00000005961
|
Adss
|
adenylosuccinate synthase |
| chr3_-_47025128 | 0.20 |
ENSRNOT00000011682
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr10_+_64930023 | 0.20 |
ENSRNOT00000071102
|
AABR07030053.1
|
|
| chr17_+_85356042 | 0.19 |
ENSRNOT00000022201
|
Commd3
|
COMM domain containing 3 |
| chr7_-_127058056 | 0.19 |
ENSRNOT00000022882
|
Cerk
|
ceramide kinase |
| chr12_-_30180115 | 0.19 |
ENSRNOT00000001202
|
Crcp
|
CGRP receptor component |
| chr8_-_13513337 | 0.19 |
ENSRNOT00000071532
|
LOC108348070
|
lysine-specific demethylase 4D |
| chr2_+_212257225 | 0.19 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr5_-_59149625 | 0.19 |
ENSRNOT00000040641
ENSRNOT00000084216 |
Spag8
|
sperm associated antigen 8 |
| chr7_-_12519154 | 0.19 |
ENSRNOT00000093376
ENSRNOT00000077681 |
Gpx4
|
glutathione peroxidase 4 |
| chr2_+_150211898 | 0.19 |
ENSRNOT00000018767
|
Sucnr1
|
succinate receptor 1 |
| chr3_+_61756148 | 0.19 |
ENSRNOT00000002134
|
Mtx2
|
metaxin 2 |
| chr10_-_56429748 | 0.19 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chr1_-_263269762 | 0.19 |
ENSRNOT00000022309
|
Got1
|
glutamic-oxaloacetic transaminase 1 |
| chr13_-_51183269 | 0.19 |
ENSRNOT00000039540
|
Ppfia4
|
PTPRF interacting protein alpha 4 |
| chr14_+_17615968 | 0.18 |
ENSRNOT00000003470
|
Rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr10_-_97582188 | 0.18 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr6_-_93562314 | 0.18 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr7_+_12229379 | 0.18 |
ENSRNOT00000060811
|
Adamtsl5
|
ADAMTS-like 5 |
| chr10_+_10967658 | 0.18 |
ENSRNOT00000004999
|
Cdip1
|
cell death-inducing p53 target 1 |
| chr5_+_157423213 | 0.18 |
ENSRNOT00000023431
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr5_-_128333805 | 0.18 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr10_-_88677055 | 0.18 |
ENSRNOT00000025590
|
Ghdc
|
GH3 domain containing |
| chr5_-_2803855 | 0.18 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr15_+_7871497 | 0.18 |
ENSRNOT00000046879
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr7_-_12432130 | 0.18 |
ENSRNOT00000077301
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr3_-_103857158 | 0.17 |
ENSRNOT00000007612
|
Emc4
|
ER membrane protein complex subunit 4 |
| chrX_+_63542191 | 0.17 |
ENSRNOT00000073955
|
Apoo
|
apolipoprotein O |
| chr9_+_116652530 | 0.17 |
ENSRNOT00000029210
|
L3mbtl4
|
l(3)mbt-like 4 (Drosophila) |
| chr8_-_87213627 | 0.17 |
ENSRNOT00000066084
|
Cox7a2
|
cytochrome c oxidase subunit 7A2 |
| chr10_-_84915471 | 0.17 |
ENSRNOT00000087322
|
Sp2
|
Sp2 transcription factor |
| chr1_+_85460888 | 0.17 |
ENSRNOT00000093384
|
Supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrb_Esrra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.5 | 1.6 | GO:0060066 | oviduct development(GO:0060066) |
| 0.4 | 1.6 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.4 | 1.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.3 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 0.9 | GO:0003331 | ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of renal output by angiotensin(GO:0002019) angiotensin-mediated drinking behavior(GO:0003051) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) positive regulation of gap junction assembly(GO:1903598) |
| 0.3 | 0.8 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.6 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.4 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 1.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.5 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 1.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.3 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.5 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.4 | GO:0002339 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.3 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.3 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.1 | GO:0097214 | regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 0.1 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.2 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 0.1 | 1.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.3 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.2 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.2 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 1.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.2 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.5 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0046133 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:1905242 | slow-twitch skeletal muscle fiber contraction(GO:0031444) response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) |
| 0.0 | 0.1 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0072138 | cell proliferation involved in mesonephros development(GO:0061209) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.6 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.8 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.1 | GO:0009751 | response to salicylic acid(GO:0009751) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:0098705 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0032912 | endodermal cell fate determination(GO:0007493) negative regulation of transforming growth factor beta1 production(GO:0032911) negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.5 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.2 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0061198 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.0 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.1 | GO:2001271 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 1.0 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 3.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.5 | GO:0045273 | respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.3 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 3.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 5.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:1903767 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 2.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.2 | 5.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.5 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.2 | 0.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 3.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.3 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.3 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0001635 | calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0003921 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.9 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0050145 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 1.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 1.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 7.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 5.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |