Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
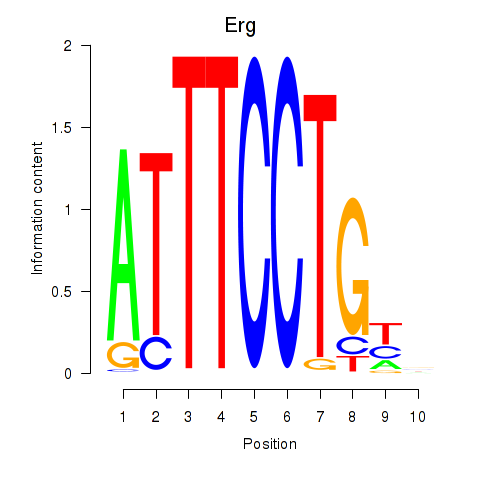
Results for Erg
Z-value: 0.66
Transcription factors associated with Erg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Erg
|
ENSRNOG00000001652 | ERG, ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Erg | rn6_v1_chr11_-_35697072_35697072 | 0.85 | 6.6e-02 | Click! |
Activity profile of Erg motif
Sorted Z-values of Erg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_190003223 | 0.81 |
ENSRNOT00000015712
|
S100a5
|
S100 calcium binding protein A5 |
| chr1_-_83993270 | 0.41 |
ENSRNOT00000002050
|
Mia
|
melanoma inhibitory activity |
| chr10_-_64642292 | 0.37 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr12_+_38144855 | 0.30 |
ENSRNOT00000032274
|
Hcar1
|
hydroxycarboxylic acid receptor 1 |
| chrX_-_115175299 | 0.30 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr1_+_255801694 | 0.24 |
ENSRNOT00000018878
|
AC103056.1
|
|
| chr2_-_190100276 | 0.23 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr7_-_36408588 | 0.22 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_-_143966863 | 0.22 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr11_+_84745904 | 0.21 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chrX_-_73778595 | 0.20 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr5_-_59085676 | 0.20 |
ENSRNOT00000068105
|
Msmp
|
microseminoprotein, prostate associated |
| chr8_-_128754514 | 0.19 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chrX_+_82143789 | 0.18 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr10_-_32471454 | 0.18 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr1_+_166141954 | 0.17 |
ENSRNOT00000042524
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr11_-_71368440 | 0.17 |
ENSRNOT00000085568
ENSRNOT00000084768 |
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr2_+_242634399 | 0.17 |
ENSRNOT00000035700
|
Emcn
|
endomucin |
| chr7_-_93826665 | 0.17 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr10_+_77537340 | 0.17 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr11_+_61531416 | 0.17 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr7_-_143967484 | 0.17 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr3_+_11679530 | 0.16 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr1_+_44311513 | 0.15 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_-_79222687 | 0.15 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr9_-_30844199 | 0.15 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr11_-_66759402 | 0.15 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr3_-_14395362 | 0.14 |
ENSRNOT00000092787
|
Rab14
|
RAB14, member RAS oncogene family |
| chr8_+_33239139 | 0.14 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_-_139907640 | 0.14 |
ENSRNOT00000045473
|
Zfp641
|
zinc finger protein 641 |
| chr1_-_198316882 | 0.14 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr19_-_22194740 | 0.14 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr15_-_42898150 | 0.13 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr18_-_6082997 | 0.13 |
ENSRNOT00000086335
ENSRNOT00000032454 |
Ss18
|
SS18, nBAF chromatin remodeling complex subunit |
| chrX_+_134979646 | 0.13 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr5_+_144031402 | 0.13 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr16_-_21362955 | 0.13 |
ENSRNOT00000039607
|
Gmip
|
Gem-interacting protein |
| chr3_+_79918969 | 0.13 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr7_-_140483693 | 0.12 |
ENSRNOT00000089060
|
Ddn
|
dendrin |
| chr1_+_224882439 | 0.12 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr14_+_70780623 | 0.12 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr7_+_58366192 | 0.12 |
ENSRNOT00000081891
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr9_+_2190915 | 0.12 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr10_-_14443010 | 0.12 |
ENSRNOT00000022142
|
Tmem204
|
transmembrane protein 204 |
| chr1_+_261802223 | 0.12 |
ENSRNOT00000081765
|
R3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr20_+_5646097 | 0.11 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr5_-_135472116 | 0.11 |
ENSRNOT00000022170
|
Nasp
|
nuclear autoantigenic sperm protein |
| chr2_-_21698937 | 0.11 |
ENSRNOT00000080165
|
AABR07007642.1
|
|
| chr2_+_41157823 | 0.11 |
ENSRNOT00000066384
|
Pde4d
|
phosphodiesterase 4D |
| chr5_-_137321121 | 0.11 |
ENSRNOT00000027414
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr16_-_74710704 | 0.11 |
ENSRNOT00000016859
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chr8_+_133210473 | 0.11 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr2_+_136993208 | 0.11 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr1_-_14412807 | 0.11 |
ENSRNOT00000074583
|
Tnfaip3
|
TNF alpha induced protein 3 |
| chr12_-_51341663 | 0.11 |
ENSRNOT00000066892
ENSRNOT00000052202 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr10_+_75088422 | 0.11 |
ENSRNOT00000081951
|
Mpo
|
myeloperoxidase |
| chr3_-_162579201 | 0.11 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr5_+_144160108 | 0.11 |
ENSRNOT00000064972
|
Eva1b
|
eva-1 homolog B |
| chr3_+_6211789 | 0.11 |
ENSRNOT00000012892
|
Rxra
|
retinoid X receptor alpha |
| chr20_+_7767395 | 0.11 |
ENSRNOT00000084771
|
Zfp523
|
zinc finger protein 523 |
| chr9_-_116222374 | 0.11 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr9_-_65736852 | 0.11 |
ENSRNOT00000087458
|
Trak2
|
trafficking kinesin protein 2 |
| chr2_-_198002625 | 0.11 |
ENSRNOT00000091888
|
BC028528
|
cDNA sequence BC028528 |
| chr3_-_113438801 | 0.11 |
ENSRNOT00000091665
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chr5_-_147635789 | 0.10 |
ENSRNOT00000037106
|
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chrX_+_1311121 | 0.10 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr7_+_28412198 | 0.10 |
ENSRNOT00000081822
ENSRNOT00000038780 ENSRNOT00000005995 |
Igf1
|
insulin-like growth factor 1 |
| chr7_-_52404774 | 0.10 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr10_+_75087892 | 0.10 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chr6_-_135829953 | 0.10 |
ENSRNOT00000080623
ENSRNOT00000039059 |
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr9_+_2202511 | 0.10 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr3_+_122836446 | 0.10 |
ENSRNOT00000028816
|
Ebf4
|
early B-cell factor 4 |
| chr13_+_48790767 | 0.10 |
ENSRNOT00000087504
|
Elk4
|
ELK4, ETS transcription factor |
| chr6_+_119519714 | 0.10 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr20_+_3363737 | 0.10 |
ENSRNOT00000079654
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr6_+_50528823 | 0.09 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr11_+_67082193 | 0.09 |
ENSRNOT00000003129
|
Cd86
|
CD86 molecule |
| chr10_-_14299167 | 0.09 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr7_-_122403667 | 0.09 |
ENSRNOT00000088814
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr17_+_15749978 | 0.09 |
ENSRNOT00000067311
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr10_+_15538872 | 0.09 |
ENSRNOT00000060078
|
Luc7l
|
LUC7-like |
| chr1_-_175420106 | 0.09 |
ENSRNOT00000013126
ENSRNOT00000077125 |
Sbf2
|
SET binding factor 2 |
| chr4_-_10020022 | 0.09 |
ENSRNOT00000044020
|
Armc10
|
armadillo repeat containing 10 |
| chr3_+_41019898 | 0.09 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr3_+_48096954 | 0.09 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr7_-_82687130 | 0.09 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr13_+_47602692 | 0.09 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr3_+_161272385 | 0.09 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr13_-_72367980 | 0.08 |
ENSRNOT00000003928
|
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr17_-_9792007 | 0.08 |
ENSRNOT00000021596
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr2_-_118882562 | 0.08 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr8_-_21968415 | 0.08 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr3_+_122114108 | 0.08 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chrX_-_123515720 | 0.08 |
ENSRNOT00000092343
|
Nkrf
|
NFKB repressing factor |
| chr2_-_185303610 | 0.08 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr13_-_88536728 | 0.08 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr10_-_85084850 | 0.08 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr7_-_70452675 | 0.08 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr5_+_64294321 | 0.08 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr14_-_82171480 | 0.08 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr2_+_188141350 | 0.08 |
ENSRNOT00000078913
|
Gon4l
|
gon-4 like |
| chr1_-_43638161 | 0.08 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_-_275876329 | 0.08 |
ENSRNOT00000047903
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr17_-_9791781 | 0.07 |
ENSRNOT00000090536
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr15_+_60084918 | 0.07 |
ENSRNOT00000012632
|
Epsti1
|
epithelial stromal interaction 1 |
| chr3_-_112676556 | 0.07 |
ENSRNOT00000014664
|
Cdan1
|
codanin 1 |
| chr13_-_111917587 | 0.07 |
ENSRNOT00000007649
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr2_+_187988925 | 0.07 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr7_-_122329443 | 0.07 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr11_+_83868655 | 0.07 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr6_+_42947334 | 0.07 |
ENSRNOT00000041376
|
RGD1563157
|
similar to 60S ribosomal protein L35 |
| chr14_-_36208278 | 0.07 |
ENSRNOT00000045050
ENSRNOT00000088448 |
Fip1l1
|
factor interacting with PAPOLA and CPSF1 |
| chr6_+_134844676 | 0.07 |
ENSRNOT00000007595
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_25174456 | 0.07 |
ENSRNOT00000092830
|
Clvs2
|
clavesin 2 |
| chr1_+_81779380 | 0.07 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chr9_+_94745217 | 0.07 |
ENSRNOT00000051338
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr15_-_34469350 | 0.07 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr3_-_122946289 | 0.07 |
ENSRNOT00000055810
|
Pced1a
|
PC-esterase domain containing 1A |
| chr11_-_60882379 | 0.07 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr8_+_60760078 | 0.07 |
ENSRNOT00000063930
|
Pstpip1
|
proline-serine-threonine phosphatase-interacting protein 1 |
| chr7_+_11077411 | 0.07 |
ENSRNOT00000007117
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr5_-_168734296 | 0.07 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr3_-_150960030 | 0.07 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr1_+_170242846 | 0.07 |
ENSRNOT00000023751
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr13_+_99005142 | 0.07 |
ENSRNOT00000004293
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr18_+_56379890 | 0.07 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr18_-_56500941 | 0.07 |
ENSRNOT00000024704
|
Hmgxb3
|
HMG-box containing 3 |
| chr2_+_183674522 | 0.07 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr2_+_32820322 | 0.07 |
ENSRNOT00000013768
|
Cd180
|
CD180 molecule |
| chr10_-_90995982 | 0.06 |
ENSRNOT00000093266
|
Gfap
|
glial fibrillary acidic protein |
| chr3_+_101010899 | 0.06 |
ENSRNOT00000073258
|
Lin7c
|
lin-7 homolog C, crumbs cell polarity complex component |
| chr9_+_52023295 | 0.06 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr8_+_102304095 | 0.06 |
ENSRNOT00000011358
|
Slc9a9
|
solute carrier family 9 member A9 |
| chr2_-_53300404 | 0.06 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr10_-_31419235 | 0.06 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_+_47019326 | 0.06 |
ENSRNOT00000006984
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr14_-_81023682 | 0.06 |
ENSRNOT00000081570
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr18_-_37245809 | 0.06 |
ENSRNOT00000079585
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_255185629 | 0.06 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr7_+_14037620 | 0.06 |
ENSRNOT00000009606
|
Syde1
|
synapse defective Rho GTPase homolog 1 |
| chr10_-_104358253 | 0.06 |
ENSRNOT00000005942
|
Caskin2
|
cask-interacting protein 2 |
| chr8_+_4440876 | 0.06 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr17_-_78812111 | 0.06 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr19_-_11451278 | 0.06 |
ENSRNOT00000026118
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr15_+_37171052 | 0.06 |
ENSRNOT00000011684
|
Zmym2
|
zinc finger MYM-type containing 2 |
| chr13_+_89774764 | 0.06 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr11_-_35749464 | 0.06 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr10_-_92602082 | 0.05 |
ENSRNOT00000007963
|
Cdc27
|
cell division cycle 27 |
| chr12_+_48628278 | 0.05 |
ENSRNOT00000000891
|
Sart3
|
squamous cell carcinoma antigen recognized by T-cells 3 |
| chr7_-_143852119 | 0.05 |
ENSRNOT00000016801
|
Rarg
|
retinoic acid receptor, gamma |
| chr6_-_76552559 | 0.05 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr6_+_110093819 | 0.05 |
ENSRNOT00000013602
|
Gpatch2l
|
G patch domain containing 2-like |
| chr13_-_55173692 | 0.05 |
ENSRNOT00000064785
ENSRNOT00000029878 ENSRNOT00000029865 ENSRNOT00000060292 ENSRNOT00000000814 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_187993758 | 0.05 |
ENSRNOT00000027182
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr10_+_47018974 | 0.05 |
ENSRNOT00000079375
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr6_-_132958546 | 0.05 |
ENSRNOT00000041903
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr10_+_89646195 | 0.05 |
ENSRNOT00000048140
|
Dhx8
|
DEAH-box helicase 8 |
| chr4_+_163162211 | 0.05 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr10_+_94988362 | 0.05 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr16_-_7250924 | 0.05 |
ENSRNOT00000025394
|
Stab1
|
stabilin 1 |
| chr18_+_44716226 | 0.05 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr11_+_57108956 | 0.05 |
ENSRNOT00000035485
|
Cd96
|
CD96 molecule |
| chr2_+_229196616 | 0.05 |
ENSRNOT00000012773
|
Ndst4
|
N-deacetylase and N-sulfotransferase 4 |
| chr2_-_187133993 | 0.05 |
ENSRNOT00000019502
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr5_+_151211342 | 0.05 |
ENSRNOT00000067939
|
Ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr14_+_78939903 | 0.05 |
ENSRNOT00000078562
ENSRNOT00000088221 |
Man2b2
Mrfap1
|
mannosidase, alpha, class 2B, member 2 Morf4 family associated protein 1 |
| chr18_+_47740328 | 0.05 |
ENSRNOT00000025119
|
Sncaip
|
synuclein, alpha interacting protein |
| chr5_+_122019301 | 0.05 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr3_+_175982777 | 0.05 |
ENSRNOT00000082073
|
Ntsr1
|
neurotensin receptor 1 |
| chr1_+_274625224 | 0.05 |
ENSRNOT00000020155
|
Pdcd4
|
programmed cell death 4 |
| chr7_-_114380613 | 0.05 |
ENSRNOT00000011898
|
Ago2
|
argonaute 2, RISC catalytic component |
| chr13_+_113373578 | 0.04 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr10_-_34166599 | 0.04 |
ENSRNOT00000003246
|
Trim41
|
tripartite motif-containing 41 |
| chr8_-_58293102 | 0.04 |
ENSRNOT00000011913
|
Rab39a
|
RAB39A, member RAS oncogene family |
| chr8_-_87419564 | 0.04 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr10_+_57660067 | 0.04 |
ENSRNOT00000010144
|
Mis12
|
MIS12 kinetochore complex component |
| chr5_+_82587420 | 0.04 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr15_+_52451161 | 0.04 |
ENSRNOT00000018725
|
Dok2
|
docking protein 2 |
| chr1_-_257498844 | 0.04 |
ENSRNOT00000019056
|
Noc3l
|
NOC3-like DNA replication regulator |
| chr1_-_82409639 | 0.04 |
ENSRNOT00000031326
|
Erich4
|
glutamate-rich 4 |
| chr16_-_9430743 | 0.04 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr7_+_70452579 | 0.04 |
ENSRNOT00000046099
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr3_+_148837814 | 0.04 |
ENSRNOT00000091808
|
Asxl1
|
additional sex combs like 1 |
| chr1_-_215310013 | 0.04 |
ENSRNOT00000054858
|
AABR07006049.2
|
|
| chr10_+_85608544 | 0.04 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr1_+_37507276 | 0.04 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chr1_-_198794947 | 0.04 |
ENSRNOT00000024639
|
Znf768
|
zinc finger protein 768 |
| chr15_-_4022223 | 0.04 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
| chr12_-_24537313 | 0.04 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr6_+_104291071 | 0.04 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr3_-_11102515 | 0.04 |
ENSRNOT00000035580
|
Fam78a
|
family with sequence similarity 78, member A |
| chr10_-_12916784 | 0.04 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr3_+_93920013 | 0.04 |
ENSRNOT00000083527
|
Lmo2
|
LIM domain only 2 |
| chr1_+_81763614 | 0.04 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chr11_+_64601029 | 0.04 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr2_+_80131563 | 0.04 |
ENSRNOT00000068484
|
Ankh
|
ANKH inorganic pyrophosphate transport regulator |
| chr15_+_67555835 | 0.04 |
ENSRNOT00000045882
|
Pcdh17
|
protocadherin 17 |
| chr5_+_149056078 | 0.04 |
ENSRNOT00000083028
|
Laptm5
|
lysosomal protein transmembrane 5 |
| chr13_-_50761306 | 0.04 |
ENSRNOT00000076610
ENSRNOT00000004241 |
Prelp
|
proline and arginine rich end leucine rich repeat protein |
| chr7_-_144778656 | 0.04 |
ENSRNOT00000055290
|
Smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Erg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.2 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.2 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.2 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.1 | 0.2 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.2 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 0.0 | 0.1 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.2 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.1 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.0 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.0 | 0.0 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.0 | 0.0 | GO:0060938 | myofibroblast differentiation(GO:0036446) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:1904714 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0043533 | inositol hexakisphosphate binding(GO:0000822) inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0051425 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |