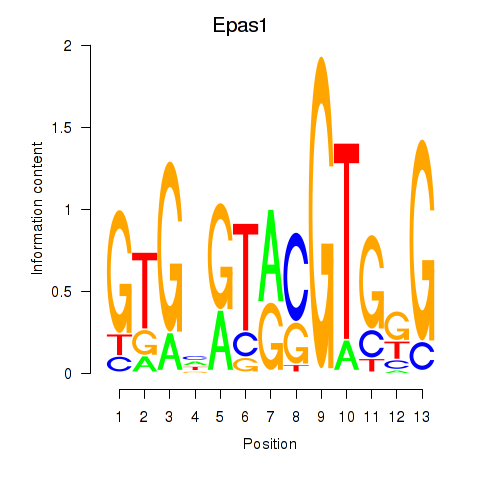
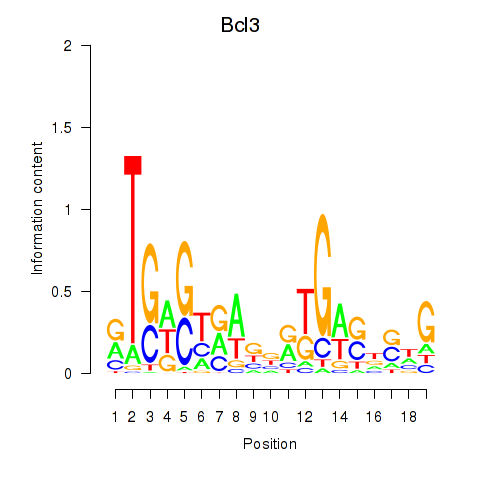
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Epas1_Bcl3
Z-value: 1.54
Transcription factors associated with Epas1_Bcl3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Epas1
|
ENSRNOG00000021318 | endothelial PAS domain protein 1 |
|
Bcl3
|
ENSRNOG00000043416 | B-cell CLL/lymphoma 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Epas1 | rn6_v1_chr6_+_10348308_10348308 | 0.84 | 7.8e-02 | Click! |
| Bcl3 | rn6_v1_chr1_-_80744831_80744831 | -0.35 | 5.6e-01 | Click! |
Activity profile of Epas1_Bcl3 motif
Sorted Z-values of Epas1_Bcl3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_245817809 | 2.56 |
ENSRNOT00000080607
|
AABR07006672.1
|
|
| chr12_-_45801842 | 1.75 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr17_+_20619324 | 1.74 |
ENSRNOT00000079788
|
AABR07027235.1
|
|
| chr3_+_71210301 | 1.50 |
ENSRNOT00000006504
|
Fam171b
|
family with sequence similarity 171, member B |
| chr2_+_30094421 | 1.30 |
ENSRNOT00000082226
|
AABR07007839.1
|
|
| chr1_+_226435979 | 1.29 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr18_-_18079560 | 1.13 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr4_-_123713319 | 1.04 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr10_+_14136883 | 1.01 |
ENSRNOT00000082295
|
AC115181.1
|
|
| chr1_+_100577056 | 0.94 |
ENSRNOT00000026992
|
Napsa
|
napsin A aspartic peptidase |
| chr1_+_104635989 | 0.85 |
ENSRNOT00000078477
|
Nav2
|
neuron navigator 2 |
| chr20_+_47779056 | 0.79 |
ENSRNOT00000090015
|
AC132752.1
|
|
| chr20_+_3558827 | 0.76 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_-_122572925 | 0.76 |
ENSRNOT00000090869
|
AC123293.2
|
|
| chr7_-_76035096 | 0.75 |
ENSRNOT00000072255
|
AABR07057530.1
|
|
| chr12_+_31934343 | 0.68 |
ENSRNOT00000011181
|
Tmem132d
|
transmembrane protein 132D |
| chr1_+_277459200 | 0.66 |
ENSRNOT00000086008
|
AABR07007023.1
|
|
| chr1_-_260992291 | 0.63 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr18_+_56193978 | 0.62 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr8_-_123829749 | 0.59 |
ENSRNOT00000090467
|
AABR07071598.1
|
|
| chr5_+_63781801 | 0.58 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr3_+_161413298 | 0.55 |
ENSRNOT00000023965
ENSRNOT00000088776 |
Mmp9
|
matrix metallopeptidase 9 |
| chrX_+_151103576 | 0.54 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chrX_-_124464963 | 0.53 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr14_+_17064353 | 0.50 |
ENSRNOT00000003052
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr5_+_16463597 | 0.49 |
ENSRNOT00000091550
|
AABR07047036.1
|
|
| chr4_-_144869919 | 0.49 |
ENSRNOT00000009066
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr8_-_94563760 | 0.47 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
| chr7_+_45328105 | 0.45 |
ENSRNOT00000029208
ENSRNOT00000090641 |
Slc6a15
|
solute carrier family 6 member 15 |
| chr3_+_110442637 | 0.45 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr9_-_20528879 | 0.45 |
ENSRNOT00000085293
|
AABR07066871.3
|
|
| chr7_+_70946228 | 0.45 |
ENSRNOT00000039306
|
Stat6
|
signal transducer and activator of transcription 6 |
| chr2_+_2694480 | 0.43 |
ENSRNOT00000083565
|
AABR07072671.1
|
|
| chr20_+_46428124 | 0.43 |
ENSRNOT00000000327
|
Foxo3
|
forkhead box O3 |
| chr1_-_255171691 | 0.43 |
ENSRNOT00000090224
|
AC113773.2
|
|
| chr7_-_3074359 | 0.41 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr5_-_113939127 | 0.40 |
ENSRNOT00000039554
|
Mysm1
|
myb-like, SWIRM and MPN domains 1 |
| chr6_+_135610743 | 0.39 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr8_+_48805684 | 0.38 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr14_-_12387102 | 0.38 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr8_-_48597867 | 0.37 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chr1_-_163554839 | 0.36 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr17_-_1085885 | 0.36 |
ENSRNOT00000026287
|
Ptch1
|
patched 1 |
| chr4_+_100209951 | 0.36 |
ENSRNOT00000015807
|
LOC691113
|
hypothetical protein LOC691113 |
| chr4_+_161720501 | 0.35 |
ENSRNOT00000008196
ENSRNOT00000055886 |
Nrip2
|
nuclear receptor interacting protein 2 |
| chr11_-_84008946 | 0.35 |
ENSRNOT00000037612
|
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr2_-_179704629 | 0.34 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr3_-_94419048 | 0.34 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr15_-_100348760 | 0.34 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr7_-_118396728 | 0.33 |
ENSRNOT00000066431
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr1_+_165625350 | 0.33 |
ENSRNOT00000087534
|
Rab6a
|
RAB6A, member RAS oncogene family |
| chr4_-_125958112 | 0.33 |
ENSRNOT00000071315
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_-_85517683 | 0.32 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr3_+_97723901 | 0.31 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr16_+_65256751 | 0.31 |
ENSRNOT00000092012
|
AABR07026137.2
|
|
| chr13_-_35668968 | 0.31 |
ENSRNOT00000042862
ENSRNOT00000003428 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_+_27973681 | 0.30 |
ENSRNOT00000004970
|
Gabrb2
|
gamma-aminobutyric acid type A receptor beta 2 subunit |
| chr9_+_73529612 | 0.30 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr17_-_77687456 | 0.29 |
ENSRNOT00000045765
ENSRNOT00000081645 |
Frmd4a
|
FERM domain containing 4A |
| chr4_-_117153907 | 0.29 |
ENSRNOT00000091374
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr18_+_60392376 | 0.29 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_+_146784915 | 0.28 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr3_+_171832500 | 0.28 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chrX_-_142131545 | 0.27 |
ENSRNOT00000077402
|
Fgf13
|
fibroblast growth factor 13 |
| chr10_+_83231238 | 0.26 |
ENSRNOT00000077625
|
Spop
|
speckle type BTB/POZ protein |
| chr10_-_90994437 | 0.26 |
ENSRNOT00000093167
|
Gfap
|
glial fibrillary acidic protein |
| chr5_-_7941822 | 0.26 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr14_+_104250617 | 0.26 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr8_-_48582353 | 0.25 |
ENSRNOT00000011582
|
Pdzd3
|
PDZ domain containing 3 |
| chr11_+_57430166 | 0.25 |
ENSRNOT00000093201
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_-_11808977 | 0.24 |
ENSRNOT00000071795
|
AABR07035375.1
|
|
| chr20_-_31956649 | 0.24 |
ENSRNOT00000072429
|
Hk1
|
hexokinase 1 |
| chr16_+_23668595 | 0.24 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_-_21492801 | 0.24 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr10_+_86157608 | 0.24 |
ENSRNOT00000082668
ENSRNOT00000008256 |
Cdk12
|
cyclin-dependent kinase 12 |
| chr13_-_67206688 | 0.24 |
ENSRNOT00000003630
ENSRNOT00000090693 |
Pla2g4a
|
phospholipase A2 group IVA |
| chr10_-_85084850 | 0.24 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr7_+_97559841 | 0.24 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr4_+_133286114 | 0.22 |
ENSRNOT00000084158
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2 |
| chr15_-_42898150 | 0.22 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr2_+_205783252 | 0.21 |
ENSRNOT00000025633
ENSRNOT00000025600 |
Trim33
|
tripartite motif-containing 33 |
| chr16_+_4469468 | 0.21 |
ENSRNOT00000021164
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr6_+_21708487 | 0.21 |
ENSRNOT00000087358
|
AABR07063197.1
|
|
| chr1_+_219250265 | 0.21 |
ENSRNOT00000024353
|
Doc2g
|
double C2-like domains, gamma |
| chr3_-_94418711 | 0.20 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr13_+_90260783 | 0.20 |
ENSRNOT00000050547
|
Cd84
|
CD84 molecule |
| chr16_-_930527 | 0.20 |
ENSRNOT00000014787
|
Spin1
|
spindlin 1 |
| chr6_-_100011226 | 0.20 |
ENSRNOT00000083286
ENSRNOT00000041442 |
Max
|
MYC associated factor X |
| chr5_-_147412705 | 0.20 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr2_-_96668222 | 0.20 |
ENSRNOT00000016567
|
Pkia
|
cAMP-dependent protein kinase inhibitor alpha |
| chr10_-_40296470 | 0.20 |
ENSRNOT00000086456
|
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr10_-_88389347 | 0.19 |
ENSRNOT00000022695
|
Klhl11
|
kelch-like family member 11 |
| chr18_-_37819097 | 0.19 |
ENSRNOT00000025880
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr15_-_13228607 | 0.19 |
ENSRNOT00000042010
ENSRNOT00000088214 |
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr10_+_84979717 | 0.18 |
ENSRNOT00000065555
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr8_-_84522588 | 0.17 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr1_+_192379543 | 0.17 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr12_+_24978483 | 0.17 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr18_+_56364620 | 0.17 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr15_+_102164751 | 0.16 |
ENSRNOT00000076400
|
Gpc6
|
glypican 6 |
| chr13_-_89242443 | 0.16 |
ENSRNOT00000029202
|
Atf6
|
activating transcription factor 6 |
| chr15_-_52317219 | 0.16 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr4_+_22445414 | 0.16 |
ENSRNOT00000087657
ENSRNOT00000030224 |
Rundc3b
|
RUN domain containing 3B |
| chr9_+_50892605 | 0.16 |
ENSRNOT00000033133
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr8_+_27777179 | 0.15 |
ENSRNOT00000009825
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 |
| chr9_-_81864202 | 0.15 |
ENSRNOT00000084964
|
Zfp142
|
zinc finger protein 142 |
| chr17_-_51912496 | 0.15 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr15_-_14737704 | 0.14 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr6_-_111015598 | 0.14 |
ENSRNOT00000015046
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr2_-_210550490 | 0.14 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr2_-_188718704 | 0.14 |
ENSRNOT00000028010
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr2_+_58448917 | 0.14 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr18_+_59748444 | 0.14 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr1_-_188895223 | 0.14 |
ENSRNOT00000032796
|
Gpr139
|
G protein-coupled receptor 139 |
| chr16_-_85306366 | 0.14 |
ENSRNOT00000089650
|
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr8_+_62482140 | 0.13 |
ENSRNOT00000026492
|
Edc3
|
enhancer of mRNA decapping 3 |
| chr3_+_55094637 | 0.13 |
ENSRNOT00000058763
|
Cers6
|
ceramide synthase 6 |
| chr1_-_129776276 | 0.13 |
ENSRNOT00000051402
|
Arrdc4
|
arrestin domain containing 4 |
| chr7_-_140454268 | 0.13 |
ENSRNOT00000081468
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr4_-_129846642 | 0.13 |
ENSRNOT00000010717
|
Frmd4b
|
FERM domain containing 4B |
| chr16_-_73670482 | 0.13 |
ENSRNOT00000024532
|
Nkx6-3
|
NK6 homeobox 3 |
| chr2_+_238257031 | 0.13 |
ENSRNOT00000016066
|
Ints12
|
integrator complex subunit 12 |
| chr13_-_49313940 | 0.13 |
ENSRNOT00000012190
|
Cntn2
|
contactin 2 |
| chr14_-_5033941 | 0.13 |
ENSRNOT00000072818
ENSRNOT00000082331 |
Zfp326
|
zinc finger protein 326 |
| chr3_+_4106477 | 0.13 |
ENSRNOT00000078985
|
AABR07051251.1
|
|
| chr14_+_15059387 | 0.12 |
ENSRNOT00000002843
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr10_-_89700283 | 0.12 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr9_+_73334618 | 0.12 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr2_+_198721724 | 0.12 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr5_+_135735825 | 0.12 |
ENSRNOT00000068267
|
Zswim5
|
zinc finger, SWIM-type containing 5 |
| chr15_-_33333417 | 0.12 |
ENSRNOT00000018982
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr1_-_7064870 | 0.11 |
ENSRNOT00000019983
|
Stx11
|
syntaxin 11 |
| chr1_-_59732409 | 0.11 |
ENSRNOT00000014824
|
Has1
|
hyaluronan synthase 1 |
| chr8_-_45137893 | 0.11 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr1_+_78767911 | 0.11 |
ENSRNOT00000022360
|
Prkd2
|
protein kinase D2 |
| chr11_+_73936750 | 0.11 |
ENSRNOT00000002350
|
Atp13a3
|
ATPase 13A3 |
| chr5_-_137125737 | 0.11 |
ENSRNOT00000088532
|
AABR07049768.1
|
|
| chr8_+_117068582 | 0.11 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr14_-_21127868 | 0.11 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr8_-_7426611 | 0.11 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr1_+_192613372 | 0.11 |
ENSRNOT00000016632
|
Cacng3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr1_+_170262156 | 0.11 |
ENSRNOT00000024077
ENSRNOT00000085395 |
Cckbr
|
cholecystokinin B receptor |
| chr7_-_62972084 | 0.11 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr1_+_190671696 | 0.11 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr17_-_29895386 | 0.10 |
ENSRNOT00000048757
|
Cdyl
|
chromodomain Y-like |
| chr1_+_164502389 | 0.10 |
ENSRNOT00000043554
|
Arrb1
|
arrestin, beta 1 |
| chr9_+_15309127 | 0.10 |
ENSRNOT00000019177
|
Prickle4
|
prickle planar cell polarity protein 4 |
| chr12_+_21721837 | 0.10 |
ENSRNOT00000073353
|
Mepce
|
methylphosphate capping enzyme |
| chr2_+_18354542 | 0.10 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr11_-_87628502 | 0.10 |
ENSRNOT00000002568
|
Med15
|
mediator complex subunit 15 |
| chr17_+_36690249 | 0.10 |
ENSRNOT00000089476
ENSRNOT00000082140 |
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr7_+_25808419 | 0.10 |
ENSRNOT00000091571
|
Rfx4
|
regulatory factor X4 |
| chr18_+_64008048 | 0.10 |
ENSRNOT00000083414
|
Ldlrad4
|
low density lipoprotein receptor class A domain containing 4 |
| chr1_+_266333440 | 0.10 |
ENSRNOT00000071243
|
Sfxn2
|
sideroflexin 2 |
| chr1_+_80920747 | 0.10 |
ENSRNOT00000026077
|
Zfp180
|
zinc finger protein 180 |
| chr13_+_60619309 | 0.09 |
ENSRNOT00000082129
|
AABR07021204.1
|
|
| chr12_+_25093524 | 0.09 |
ENSRNOT00000081084
|
Eif4h
|
eukaryotic translation initiation factor 4H |
| chr18_+_17043903 | 0.09 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr4_-_180234804 | 0.09 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr4_-_157372861 | 0.09 |
ENSRNOT00000021578
|
P3h3
|
prolyl 3-hydroxylase 3 |
| chr6_-_39363367 | 0.09 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr9_-_44419998 | 0.09 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr5_-_7874909 | 0.08 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_84328114 | 0.08 |
ENSRNOT00000028266
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr1_+_176854816 | 0.07 |
ENSRNOT00000087461
|
Usp47
|
ubiquitin specific peptidase 47 |
| chr10_+_17261541 | 0.07 |
ENSRNOT00000005478
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr5_+_31568419 | 0.07 |
ENSRNOT00000050137
|
Mmp16
|
matrix metallopeptidase 16 |
| chr1_+_82174451 | 0.07 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chr2_+_178679395 | 0.07 |
ENSRNOT00000088686
|
Fam198b
|
family with sequence similarity 198, member B |
| chr12_-_38691075 | 0.07 |
ENSRNOT00000084192
|
Bcl7a
|
BCL tumor suppressor 7A |
| chr1_+_177183209 | 0.07 |
ENSRNOT00000085326
|
Micalcl
|
MICAL C-terminal like |
| chr8_+_408001 | 0.07 |
ENSRNOT00000046058
|
Gucy1a2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr14_-_20920286 | 0.06 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr3_-_52510507 | 0.06 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr1_-_265798167 | 0.06 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr9_+_81783349 | 0.06 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr10_-_63491713 | 0.06 |
ENSRNOT00000063941
|
Tusc5
|
tumor suppressor candidate 5 |
| chr8_+_111495331 | 0.06 |
ENSRNOT00000091275
ENSRNOT00000012162 |
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr1_+_89491654 | 0.06 |
ENSRNOT00000028632
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr12_+_48598647 | 0.06 |
ENSRNOT00000000889
|
Tmem119
|
transmembrane protein 119 |
| chr20_+_44803666 | 0.06 |
ENSRNOT00000057227
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr6_-_107678156 | 0.06 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr1_+_80953275 | 0.06 |
ENSRNOT00000087253
|
Zfp112
|
zinc finger protein 112 |
| chr15_-_84748525 | 0.06 |
ENSRNOT00000090009
|
Klf12
|
Kruppel-like factor 12 |
| chr5_+_137356801 | 0.06 |
ENSRNOT00000027432
|
RGD1305347
|
similar to RIKEN cDNA 2610528J11 |
| chr1_+_140792049 | 0.05 |
ENSRNOT00000049681
|
Acan
|
aggrecan |
| chr17_-_9792007 | 0.05 |
ENSRNOT00000021596
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr12_+_25498198 | 0.05 |
ENSRNOT00000076916
|
Ncf1
|
neutrophil cytosolic factor 1 |
| chr11_-_27094364 | 0.05 |
ENSRNOT00000050902
|
Rwdd2b
|
RWD domain containing 2B |
| chr2_+_62150493 | 0.05 |
ENSRNOT00000080241
|
Zfr
|
zinc finger RNA binding protein |
| chr20_+_44803885 | 0.05 |
ENSRNOT00000077936
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr5_-_62186372 | 0.05 |
ENSRNOT00000089789
|
Coro2a
|
coronin 2A |
| chr17_-_6244612 | 0.05 |
ENSRNOT00000042145
ENSRNOT00000090914 ENSRNOT00000082611 |
Ntrk2
|
neurotrophic receptor tyrosine kinase 2 |
| chr2_+_211546560 | 0.04 |
ENSRNOT00000033443
|
Aknad1
|
AKNA domain containing 1 |
| chr16_-_755990 | 0.04 |
ENSRNOT00000013501
|
Polr3a
|
RNA polymerase III subunit A |
| chr9_-_60923706 | 0.04 |
ENSRNOT00000047517
|
AABR07067749.1
|
|
| chr13_-_70321722 | 0.04 |
ENSRNOT00000092397
ENSRNOT00000092336 ENSRNOT00000092649 ENSRNOT00000092269 |
Smg7
|
SMG7 nonsense mediated mRNA decay factor |
| chr14_+_99529284 | 0.04 |
ENSRNOT00000006897
ENSRNOT00000067134 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr15_+_28023018 | 0.04 |
ENSRNOT00000090272
|
Rnase4
|
ribonuclease A family member 4 |
| chr1_-_72727112 | 0.04 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Epas1_Bcl3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 0.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.5 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to iron(III) ion(GO:0071283) |
| 0.2 | 0.6 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.2 | 0.5 | GO:0015820 | leucine transport(GO:0015820) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.6 | GO:1900623 | regulation of mast cell cytokine production(GO:0032763) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.6 | GO:0098989 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) NMDA selective glutamate receptor signaling pathway(GO:0098989) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.1 | 0.3 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:0042640 | anagen(GO:0042640) regulation of anagen(GO:0051884) |
| 0.1 | 1.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.4 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.3 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) |
| 0.1 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.2 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.1 | 0.3 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.1 | 0.2 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.2 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.0 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.3 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 1.0 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.3 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.2 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0098830 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.2 | 0.6 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 1.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.1 | 0.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030977 | taurine binding(GO:0030977) |
| 0.1 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |