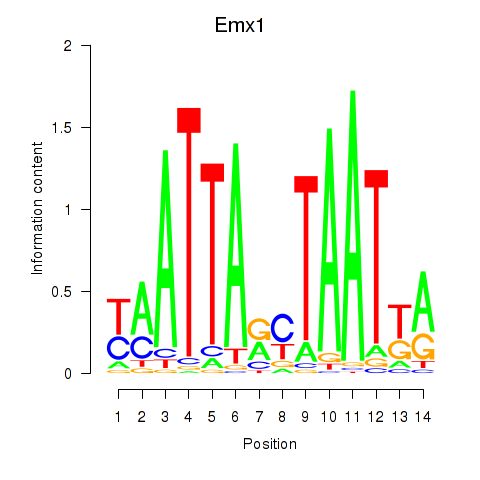
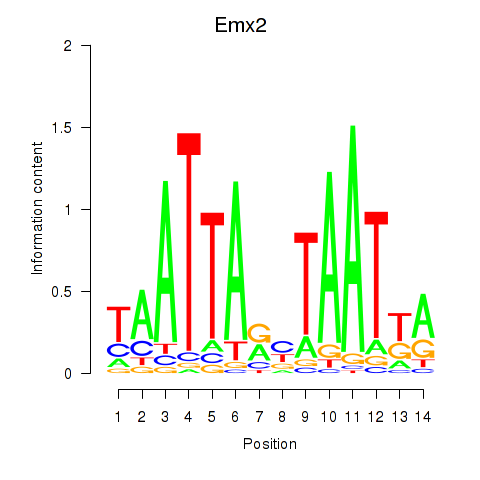
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Emx1_Emx2
Z-value: 0.29
Transcription factors associated with Emx1_Emx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Emx1
|
ENSRNOG00000015493 | empty spiracles homeobox 1 |
|
Emx2
|
ENSRNOG00000009482 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Emx2 | rn6_v1_chr1_+_280633938_280633938 | 0.90 | 4.0e-02 | Click! |
| Emx1 | rn6_v1_chr4_+_116968000_116968000 | 0.56 | 3.2e-01 | Click! |
Activity profile of Emx1_Emx2 motif
Sorted Z-values of Emx1_Emx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_73378057 | 0.11 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr20_+_3176107 | 0.11 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr2_+_86996798 | 0.09 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr11_-_11585078 | 0.09 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr13_-_76049363 | 0.08 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr2_+_86996497 | 0.08 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr17_-_51912496 | 0.07 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr20_+_34258791 | 0.06 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr12_-_46493203 | 0.06 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr9_-_30844199 | 0.06 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr1_+_101603222 | 0.06 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr20_-_13994794 | 0.05 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr2_+_54466280 | 0.05 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr3_+_161236898 | 0.05 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr20_+_3246739 | 0.05 |
ENSRNOT00000061299
|
RT1-T24-2
|
RT1 class I, locus T24, gene 2 |
| chrX_+_78042859 | 0.05 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr3_+_15560712 | 0.05 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_+_64769089 | 0.05 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_198706428 | 0.05 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr11_-_782954 | 0.05 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr11_+_40509078 | 0.04 |
ENSRNOT00000023108
|
AABR07033851.1
|
|
| chr6_+_106084815 | 0.04 |
ENSRNOT00000058195
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr13_-_32427177 | 0.04 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr8_-_3272306 | 0.04 |
ENSRNOT00000040365
|
AABR07068955.1
|
|
| chrX_+_84064427 | 0.04 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr11_-_768644 | 0.04 |
ENSRNOT00000090197
|
Epha3
|
Eph receptor A3 |
| chr13_-_67609469 | 0.03 |
ENSRNOT00000059984
ENSRNOT00000059982 |
RGD1307830
|
similar to cDNA sequence BC003331 |
| chr10_-_91661558 | 0.03 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr15_+_12827707 | 0.03 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr5_+_152680407 | 0.03 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr1_+_170205591 | 0.03 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr1_+_265157379 | 0.03 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr10_-_36402619 | 0.03 |
ENSRNOT00000040346
|
Zfp2
|
zinc finger protein 2 |
| chr20_+_3230052 | 0.03 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr1_-_189911571 | 0.03 |
ENSRNOT00000080996
ENSRNOT00000088536 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr5_+_50381244 | 0.03 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr3_+_148327965 | 0.03 |
ENSRNOT00000010851
ENSRNOT00000088481 |
Tpx2
|
TPX2, microtubule nucleation factor |
| chr1_+_219403970 | 0.03 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr4_+_31229913 | 0.03 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr1_+_189233141 | 0.03 |
ENSRNOT00000049380
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr3_-_48831417 | 0.03 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr10_+_1920529 | 0.03 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr1_+_141218095 | 0.03 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr8_+_117170620 | 0.03 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr2_+_58448917 | 0.03 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr4_-_55011415 | 0.03 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr13_-_102857551 | 0.03 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr16_+_39827749 | 0.03 |
ENSRNOT00000074885
|
Wdr17
|
WD repeat domain 17 |
| chr6_-_3254779 | 0.03 |
ENSRNOT00000061975
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr4_+_99823252 | 0.03 |
ENSRNOT00000013587
|
Polr1a
|
RNA polymerase I subunit A |
| chr1_-_166912524 | 0.03 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr16_-_32439421 | 0.03 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chr16_-_32421005 | 0.03 |
ENSRNOT00000082712
|
Nek1
|
NIMA-related kinase 1 |
| chr9_+_111279495 | 0.02 |
ENSRNOT00000072791
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr1_+_22332090 | 0.02 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chrX_-_14972675 | 0.02 |
ENSRNOT00000079664
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr2_+_52855671 | 0.02 |
ENSRNOT00000080851
|
AABR07008319.1
|
|
| chr16_+_70644474 | 0.02 |
ENSRNOT00000045955
|
LOC100359503
|
ribosomal protein S28-like |
| chr8_+_106816152 | 0.02 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr1_+_147713892 | 0.02 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr2_-_186245163 | 0.02 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr7_+_27620458 | 0.02 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr2_-_219262901 | 0.02 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr8_+_107229832 | 0.02 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr2_-_118882562 | 0.02 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr2_-_57600820 | 0.02 |
ENSRNOT00000083247
|
Nipbl
|
NIPBL, cohesin loading factor |
| chr1_-_151106802 | 0.02 |
ENSRNOT00000021971
|
Tyr
|
tyrosinase |
| chr16_+_62153792 | 0.02 |
ENSRNOT00000074543
|
Smim18
|
small integral membrane protein 18 |
| chr17_-_18590536 | 0.02 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr3_-_55951584 | 0.02 |
ENSRNOT00000036585
|
Fastkd1
|
FAST kinase domains 1 |
| chr7_-_50278842 | 0.02 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr16_+_46731403 | 0.02 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr2_+_220432037 | 0.02 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr1_-_256734719 | 0.02 |
ENSRNOT00000021546
ENSRNOT00000089456 |
Myof
|
myoferlin |
| chr6_-_23404368 | 0.02 |
ENSRNOT00000036374
|
Fam179a
|
family with sequence similarity 179, member A |
| chr12_+_25435567 | 0.02 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr11_+_77644541 | 0.02 |
ENSRNOT00000074688
|
Tmem207
|
transmembrane protein 207 |
| chr18_+_1866080 | 0.02 |
ENSRNOT00000018646
|
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr7_-_76488216 | 0.02 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr2_+_232104809 | 0.02 |
ENSRNOT00000083193
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr2_+_24668116 | 0.02 |
ENSRNOT00000067022
ENSRNOT00000036748 |
Wdr41
|
WD repeat domain 41 |
| chrM_+_11736 | 0.02 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrX_+_131381134 | 0.02 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr5_+_145145983 | 0.02 |
ENSRNOT00000079732
|
Zmym6
|
zinc finger MYM-type containing 6 |
| chr7_-_80457816 | 0.02 |
ENSRNOT00000039430
|
AABR07057617.1
|
|
| chr13_+_57243877 | 0.02 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr2_+_18392142 | 0.02 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr15_+_52265557 | 0.02 |
ENSRNOT00000015969
|
Nudt18
|
nudix hydrolase 18 |
| chr5_+_33784715 | 0.02 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr4_+_35279063 | 0.02 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr1_+_128637049 | 0.02 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_+_137323713 | 0.02 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr9_+_94525925 | 0.02 |
ENSRNOT00000075967
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr15_-_14510828 | 0.02 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr17_-_76281660 | 0.02 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr4_+_31704747 | 0.02 |
ENSRNOT00000047850
|
AABR07059675.1
|
|
| chr4_+_173732248 | 0.02 |
ENSRNOT00000041499
|
Pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr15_-_29548400 | 0.02 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr3_-_21904133 | 0.02 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr1_-_167911961 | 0.02 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr6_-_14523961 | 0.02 |
ENSRNOT00000071402
|
Nrxn1
|
neurexin 1 |
| chr14_-_106248539 | 0.02 |
ENSRNOT00000067668
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr2_-_186245342 | 0.02 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr16_-_64806050 | 0.02 |
ENSRNOT00000015725
|
Dusp26
|
dual specificity phosphatase 26 |
| chr14_+_69800156 | 0.02 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chrX_-_139916883 | 0.02 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr13_+_105684420 | 0.02 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr20_-_45126062 | 0.02 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr7_+_91384187 | 0.02 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr2_-_250923744 | 0.02 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr14_+_108415068 | 0.02 |
ENSRNOT00000083763
|
Pus10
|
pseudouridylate synthase 10 |
| chr7_-_143497108 | 0.02 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr6_+_132551394 | 0.02 |
ENSRNOT00000019583
|
Evl
|
Enah/Vasp-like |
| chrM_+_7006 | 0.02 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr8_+_79638696 | 0.02 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr11_+_61605937 | 0.02 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
| chr6_+_91532467 | 0.02 |
ENSRNOT00000082500
ENSRNOT00000064668 |
Klhdc1
|
kelch domain containing 1 |
| chr8_-_126390801 | 0.02 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr14_+_58877806 | 0.02 |
ENSRNOT00000051559
|
RGD1562755
|
similar to 60S ribosomal protein L23a |
| chr1_-_101131413 | 0.02 |
ENSRNOT00000093729
|
Flt3lg
|
fms-related tyrosine kinase 3 ligand |
| chr2_+_88217188 | 0.02 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chr13_-_102942863 | 0.01 |
ENSRNOT00000003198
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr6_+_109617596 | 0.01 |
ENSRNOT00000085031
ENSRNOT00000089972 ENSRNOT00000082402 |
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr13_+_76942928 | 0.01 |
ENSRNOT00000040505
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr1_+_156552328 | 0.01 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr13_-_105684374 | 0.01 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr1_-_183763664 | 0.01 |
ENSRNOT00000044231
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr1_-_133503194 | 0.01 |
ENSRNOT00000077049
|
Mctp2
|
multiple C2 and transmembrane domain containing 2 |
| chr10_+_90377103 | 0.01 |
ENSRNOT00000040472
|
Grn
|
granulin precursor |
| chr12_+_41486076 | 0.01 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr1_+_21613148 | 0.01 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_-_246010594 | 0.01 |
ENSRNOT00000023762
|
Rfx3
|
regulatory factor X3 |
| chr4_+_88423412 | 0.01 |
ENSRNOT00000040163
|
Vom1r88
|
vomeronasal 1 receptor 88 |
| chr3_-_150156839 | 0.01 |
ENSRNOT00000036217
|
LOC103691939
|
elongation factor 1-alpha 1 pseudogene |
| chr18_+_70739492 | 0.01 |
ENSRNOT00000085601
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr1_+_81499821 | 0.01 |
ENSRNOT00000027100
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr6_+_129609375 | 0.01 |
ENSRNOT00000083732
|
Papola
|
poly (A) polymerase alpha |
| chr1_-_78180216 | 0.01 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr10_+_90376933 | 0.01 |
ENSRNOT00000028557
|
Grn
|
granulin precursor |
| chr10_+_45307007 | 0.01 |
ENSRNOT00000003885
|
Trim17
|
tripartite motif-containing 17 |
| chr1_-_252461461 | 0.01 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr2_+_55747318 | 0.01 |
ENSRNOT00000050655
|
Dab2
|
DAB2, clathrin adaptor protein |
| chr16_+_2537248 | 0.01 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chrX_+_9956370 | 0.01 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr5_-_162602339 | 0.01 |
ENSRNOT00000071779
|
RGD1563334
|
similar to novel protein similar to esterases |
| chr17_+_53965562 | 0.01 |
ENSRNOT00000090387
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr15_-_28044210 | 0.01 |
ENSRNOT00000033809
|
Rnase1l1
|
ribonuclease, RNase A family, 1-like 1 (pancreatic) |
| chr20_-_3166569 | 0.01 |
ENSRNOT00000091390
|
AABR07044362.1
|
|
| chr11_+_80742467 | 0.01 |
ENSRNOT00000002507
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr3_-_161246351 | 0.01 |
ENSRNOT00000020348
|
Tnnc2
|
troponin C2, fast skeletal type |
| chr2_+_240642847 | 0.01 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_-_86393141 | 0.01 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr7_-_104541392 | 0.01 |
ENSRNOT00000078116
|
Fam49b
|
family with sequence similarity 49, member B |
| chrX_-_72034099 | 0.01 |
ENSRNOT00000004310
|
Ercc6l
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr9_+_95398237 | 0.01 |
ENSRNOT00000025879
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_+_36691875 | 0.01 |
ENSRNOT00000074314
|
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr11_+_31539016 | 0.01 |
ENSRNOT00000072856
|
Ifnar2
|
interferon alpha and beta receptor subunit 2 |
| chr8_+_111790678 | 0.01 |
ENSRNOT00000013265
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr1_-_252116971 | 0.01 |
ENSRNOT00000035013
|
Lipo1
|
lipase, member O1 |
| chr7_-_144993652 | 0.01 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr3_-_104504204 | 0.01 |
ENSRNOT00000049582
|
Ryr3
|
ryanodine receptor 3 |
| chr1_+_166983175 | 0.01 |
ENSRNOT00000027028
|
Anapc15
|
anaphase promoting complex subunit 15 |
| chr2_+_166348373 | 0.01 |
ENSRNOT00000033132
|
RGD1563620
|
similar to retinoblastoma binding protein 4 |
| chr1_+_22364551 | 0.01 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr19_-_10513349 | 0.01 |
ENSRNOT00000061270
|
Adgrg5
|
adhesion G protein-coupled receptor G5 |
| chr2_+_2903492 | 0.01 |
ENSRNOT00000062017
|
Ttc37
|
tetratricopeptide repeat domain 37 |
| chr19_-_14945302 | 0.01 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr9_+_66058047 | 0.01 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr9_+_95161157 | 0.01 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chrM_+_10160 | 0.01 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_-_62690806 | 0.01 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr5_+_108278217 | 0.01 |
ENSRNOT00000038972
|
Dmrta1
|
DMRT-like family A1 |
| chr6_-_54059119 | 0.01 |
ENSRNOT00000005521
|
Hdac9
|
histone deacetylase 9 |
| chr2_-_149444548 | 0.01 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr8_+_70447040 | 0.01 |
ENSRNOT00000016798
|
Ints14
|
integrator complex subunit 14 |
| chr14_-_45165207 | 0.01 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr2_+_41442241 | 0.01 |
ENSRNOT00000067546
|
Pde4d
|
phosphodiesterase 4D |
| chr17_-_21577085 | 0.01 |
ENSRNOT00000020737
|
Tmem14c
|
transmembrane protein 14C |
| chr17_-_76250537 | 0.01 |
ENSRNOT00000092693
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr6_+_18880737 | 0.01 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_+_118831350 | 0.01 |
ENSRNOT00000083720
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr2_-_122641409 | 0.01 |
ENSRNOT00000068334
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr8_-_55144087 | 0.01 |
ENSRNOT00000039045
|
Dixdc1
|
DIX domain containing 1 |
| chr5_+_167141875 | 0.01 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr1_+_79631668 | 0.01 |
ENSRNOT00000083546
ENSRNOT00000035286 |
Mill1
|
MHC I like leukocyte 1 |
| chr10_-_88000423 | 0.01 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr1_+_61786900 | 0.01 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr15_+_34234755 | 0.01 |
ENSRNOT00000059987
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr17_+_63635086 | 0.01 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr2_+_248398917 | 0.01 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr8_-_128711221 | 0.01 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr5_+_150459713 | 0.00 |
ENSRNOT00000081681
ENSRNOT00000074251 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chr19_-_11669578 | 0.00 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr15_-_52399074 | 0.00 |
ENSRNOT00000018440
|
Xpo7
|
exportin 7 |
| chr4_+_163349125 | 0.00 |
ENSRNOT00000084823
|
Klre1
|
killer cell lectin-like receptor, family E, member 1 |
| chr5_-_164648328 | 0.00 |
ENSRNOT00000090811
|
Zfp933
|
zinc finger protein 933 |
| chr11_-_60546997 | 0.00 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr20_+_3155652 | 0.00 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr13_+_92264231 | 0.00 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr9_-_97290639 | 0.00 |
ENSRNOT00000026491
ENSRNOT00000056724 |
Iqca1
|
IQ motif containing with AAA domain 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Emx1_Emx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032759 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.0 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |