Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
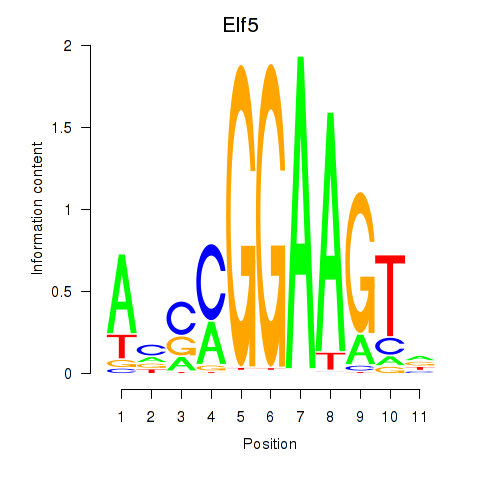
Results for Elf5
Z-value: 0.92
Transcription factors associated with Elf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Elf5
|
ENSRNOG00000008282 | E74-like factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elf5 | rn6_v1_chr3_+_93351619_93351619 | 0.13 | 8.4e-01 | Click! |
Activity profile of Elf5 motif
Sorted Z-values of Elf5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_42933077 | 0.46 |
ENSRNOT00000003361
|
Mfap3
|
microfibrillar-associated protein 3 |
| chr6_+_108796182 | 0.40 |
ENSRNOT00000006297
|
Fcf1
|
FCF1 rRNA-processing protein |
| chr20_+_5441876 | 0.34 |
ENSRNOT00000092476
|
Rps18
|
ribosomal protein S18 |
| chr10_-_10767389 | 0.33 |
ENSRNOT00000066754
|
Smim22
|
small integral membrane protein 22 |
| chr14_+_42007312 | 0.33 |
ENSRNOT00000063985
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr8_+_21663325 | 0.32 |
ENSRNOT00000027749
|
Ubl5
|
ubiquitin-like 5 |
| chr19_+_49016891 | 0.30 |
ENSRNOT00000016713
|
Dynlrb2
|
dynein light chain roadblock-type 2 |
| chr5_-_147803851 | 0.28 |
ENSRNOT00000071922
|
Eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr3_-_161294125 | 0.28 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chr1_-_143169657 | 0.28 |
ENSRNOT00000025888
|
Rps17
|
ribosomal protein S17 |
| chr3_-_111560556 | 0.28 |
ENSRNOT00000030532
|
Ltk
|
leukocyte receptor tyrosine kinase |
| chr8_-_48726963 | 0.28 |
ENSRNOT00000016145
|
Trappc4
|
trafficking protein particle complex 4 |
| chr11_-_24294179 | 0.26 |
ENSRNOT00000002116
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr4_-_34282351 | 0.25 |
ENSRNOT00000011123
|
Rpa3
|
replication protein A3 |
| chrX_-_156440461 | 0.25 |
ENSRNOT00000083951
|
Rpl10
|
ribosomal protein L10 |
| chr1_-_219438779 | 0.24 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr3_-_46726946 | 0.23 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr15_-_34352673 | 0.23 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr3_-_151688149 | 0.22 |
ENSRNOT00000072945
|
Nfs1
|
NFS1 cysteine desulfurase |
| chr10_-_30118873 | 0.22 |
ENSRNOT00000006063
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr19_+_37282018 | 0.22 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr1_-_281101438 | 0.21 |
ENSRNOT00000012734
|
Rab11fip2
|
RAB11 family interacting protein 2 |
| chr1_+_201687758 | 0.21 |
ENSRNOT00000093308
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr10_-_84976170 | 0.21 |
ENSRNOT00000013542
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr2_+_4195917 | 0.21 |
ENSRNOT00000093303
|
RGD1560883
|
similar to KIAA0825 protein |
| chr6_+_129519709 | 0.21 |
ENSRNOT00000006307
|
Gskip
|
GSK3B interacting protein |
| chr10_+_75032365 | 0.21 |
ENSRNOT00000010448
|
Supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr2_+_187447501 | 0.19 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chrX_+_20351486 | 0.19 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr13_+_51958834 | 0.19 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr11_+_87435185 | 0.19 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr10_-_16792909 | 0.19 |
ENSRNOT00000004653
|
Atp6v0e1
|
ATPase H+ transporting V0 subunit e1 |
| chr11_+_32655616 | 0.19 |
ENSRNOT00000084412
ENSRNOT00000034383 |
Clic6
|
chloride intracellular channel 6 |
| chr16_-_7588841 | 0.19 |
ENSRNOT00000084645
|
Mettl6
|
methyltransferase like 6 |
| chr17_+_9736577 | 0.19 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr5_-_16706909 | 0.19 |
ENSRNOT00000011314
|
Rps20
|
ribosomal protein S20 |
| chr1_-_198120061 | 0.18 |
ENSRNOT00000026231
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr5_+_116973159 | 0.18 |
ENSRNOT00000046475
|
LOC100912024
|
uncharacterized LOC100912024 |
| chr4_+_56744561 | 0.18 |
ENSRNOT00000009737
|
Atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr20_+_1889652 | 0.18 |
ENSRNOT00000068508
ENSRNOT00000000996 |
Olr1746
|
olfactory receptor 1746 |
| chr14_+_24129592 | 0.18 |
ENSRNOT00000040647
|
LOC689899
|
similar to 60S ribosomal protein L23a |
| chr1_+_174132798 | 0.18 |
ENSRNOT00000019247
|
Rpl27a
|
ribosomal protein L27a |
| chr15_+_34352914 | 0.18 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr3_+_140106766 | 0.18 |
ENSRNOT00000014046
|
Naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr6_+_102392828 | 0.18 |
ENSRNOT00000089162
|
Rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr3_+_175629374 | 0.18 |
ENSRNOT00000084027
|
Rps21
|
ribosomal protein S21 |
| chr10_+_94566928 | 0.17 |
ENSRNOT00000078446
|
Prr29
|
proline rich 29 |
| chr17_+_5311274 | 0.17 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr3_+_47578384 | 0.17 |
ENSRNOT00000082769
|
Psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr10_-_34166599 | 0.17 |
ENSRNOT00000003246
|
Trim41
|
tripartite motif-containing 41 |
| chr1_-_247476827 | 0.17 |
ENSRNOT00000021298
|
Insl6
|
insulin-like 6 |
| chr4_-_147719072 | 0.17 |
ENSRNOT00000014493
|
Rpl32
|
ribosomal protein L32 |
| chr8_-_116297850 | 0.17 |
ENSRNOT00000031739
ENSRNOT00000082320 |
Cyb561d2
|
cytochrome b561 family, member D2 |
| chr20_-_14020007 | 0.17 |
ENSRNOT00000093521
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr2_-_4195810 | 0.16 |
ENSRNOT00000018457
|
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr5_-_142845116 | 0.16 |
ENSRNOT00000065105
|
RGD1559909
|
RGD1559909 |
| chr2_+_187415262 | 0.16 |
ENSRNOT00000056923
|
Gpatch4
|
G patch domain containing 4 |
| chr2_-_147819335 | 0.16 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr1_-_89017290 | 0.16 |
ENSRNOT00000028438
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr1_+_197999336 | 0.16 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr8_-_127900463 | 0.16 |
ENSRNOT00000078303
|
Slc22a13
|
solute carrier family 22 member 13 |
| chr5_+_57947716 | 0.16 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chrX_-_45419817 | 0.16 |
ENSRNOT00000087509
|
Ptges3l1
|
prostaglandin E synthase 3-like 1 |
| chr13_+_83996080 | 0.15 |
ENSRNOT00000004403
ENSRNOT00000070958 |
Cd247
|
Cd247 molecule |
| chr15_-_42794279 | 0.15 |
ENSRNOT00000023385
|
Ephx2
|
epoxide hydrolase 2 |
| chr10_+_109630099 | 0.15 |
ENSRNOT00000072099
|
Ccdc137
|
coiled-coil domain containing 137 |
| chr20_+_46044892 | 0.15 |
ENSRNOT00000057187
|
Ak9
|
adenylate kinase 9 |
| chr4_-_133127175 | 0.15 |
ENSRNOT00000057758
|
Shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr8_+_48727618 | 0.15 |
ENSRNOT00000029546
|
Rps25
|
ribosomal protein s25 |
| chr16_+_3817757 | 0.15 |
ENSRNOT00000088566
|
Tmem254
|
transmembrane protein 254 |
| chr1_+_219481575 | 0.15 |
ENSRNOT00000025507
|
Pold4
|
DNA polymerase delta 4, accessory subunit |
| chr1_-_89084859 | 0.15 |
ENSRNOT00000032026
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr7_+_143929662 | 0.15 |
ENSRNOT00000017942
|
Myg1
|
melanocyte proliferating gene 1 |
| chr8_+_133029625 | 0.15 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr5_-_17061361 | 0.15 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr12_-_21923281 | 0.15 |
ENSRNOT00000075641
|
RGD1561143
|
similar to cell surface receptor FDFACT |
| chrX_+_15098904 | 0.15 |
ENSRNOT00000007367
ENSRNOT00000087033 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr2_-_27287605 | 0.15 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr17_-_21739408 | 0.15 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_-_252382955 | 0.15 |
ENSRNOT00000021705
|
Rpf1
|
ribosome production factor 1 homolog |
| chr17_+_22619891 | 0.15 |
ENSRNOT00000060403
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr14_+_115166416 | 0.15 |
ENSRNOT00000088916
ENSRNOT00000078329 |
Psme4
|
proteasome activator subunit 4 |
| chr8_-_48727154 | 0.15 |
ENSRNOT00000088471
|
Trappc4
|
trafficking protein particle complex 4 |
| chr3_+_94530586 | 0.15 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr1_-_80783898 | 0.15 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr9_-_29647903 | 0.15 |
ENSRNOT00000019062
|
Ogfrl1
|
opioid growth factor receptor-like 1 |
| chr20_-_11644166 | 0.15 |
ENSRNOT00000073009
|
LOC100361739
|
keratin associated protein 12-1-like |
| chr8_-_27852996 | 0.14 |
ENSRNOT00000037790
|
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr8_-_117820413 | 0.14 |
ENSRNOT00000075819
|
Tma7
|
translation machinery associated 7 homolog |
| chr8_+_55037750 | 0.14 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr3_+_95959703 | 0.14 |
ENSRNOT00000006406
|
Immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr12_+_19680712 | 0.14 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr7_-_140090922 | 0.14 |
ENSRNOT00000014456
|
Lalba
|
lactalbumin, alpha |
| chrX_+_105402867 | 0.14 |
ENSRNOT00000057222
|
Rpl36a
|
ribosomal protein L36a |
| chr1_+_222198534 | 0.14 |
ENSRNOT00000028712
|
Bad
|
BCL2-associated agonist of cell death |
| chr6_-_125590049 | 0.14 |
ENSRNOT00000006371
|
Tc2n
|
tandem C2 domains, nuclear |
| chr15_+_43582020 | 0.14 |
ENSRNOT00000013001
|
Pnma2
|
paraneoplastic Ma antigen 2 |
| chr5_-_154394328 | 0.14 |
ENSRNOT00000030043
|
Rpl11
|
ribosomal protein L11 |
| chr1_-_174620064 | 0.14 |
ENSRNOT00000016224
|
Tmem41b
|
transmembrane protein 41B |
| chr8_-_55491152 | 0.14 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr5_+_142845114 | 0.14 |
ENSRNOT00000039870
|
Yrdc
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr8_-_71533459 | 0.14 |
ENSRNOT00000085745
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr5_-_140024176 | 0.14 |
ENSRNOT00000016492
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr7_+_122700854 | 0.14 |
ENSRNOT00000086603
|
Rbx1
|
ring-box 1 |
| chr15_-_43667029 | 0.14 |
ENSRNOT00000014798
|
Bnip3l
|
BCL2/adenovirus E1B interacting protein 3-like |
| chr2_+_244521699 | 0.14 |
ENSRNOT00000029382
|
Stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr15_+_33333420 | 0.13 |
ENSRNOT00000049927
|
RGD1308430
|
similar to 1700123O20Rik protein |
| chr1_+_31967978 | 0.13 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr3_+_32947378 | 0.13 |
ENSRNOT00000007404
|
Acvr2a
|
activin A receptor type 2A |
| chr5_-_135663284 | 0.13 |
ENSRNOT00000087237
ENSRNOT00000066869 |
Toe1
|
target of EGR1, member 1 (nuclear) |
| chr10_+_14105750 | 0.13 |
ENSRNOT00000090552
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr9_-_92435363 | 0.13 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr10_+_55138633 | 0.13 |
ENSRNOT00000057223
|
Mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr4_-_117589464 | 0.13 |
ENSRNOT00000021167
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chr13_+_34267471 | 0.13 |
ENSRNOT00000061619
ENSRNOT00000092802 |
Nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr9_+_67699379 | 0.13 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chrX_-_21710678 | 0.13 |
ENSRNOT00000004184
|
Ribc1
|
RIB43A domain with coiled-coils 1 |
| chr8_+_55050284 | 0.13 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr6_-_102095201 | 0.13 |
ENSRNOT00000014125
|
Plek2
|
pleckstrin 2 |
| chr6_+_86651196 | 0.13 |
ENSRNOT00000034756
|
RGD1307621
|
hypothetical LOC314168 |
| chr8_+_62298358 | 0.13 |
ENSRNOT00000025525
|
Cox5a
|
cytochrome c oxidase subunit 5A |
| chr8_+_117953444 | 0.13 |
ENSRNOT00000028141
|
Cdc25a
|
cell division cycle 25A |
| chr1_-_30917882 | 0.13 |
ENSRNOT00000015440
|
Echdc1
|
ethylmalonyl-CoA decarboxylase 1 |
| chr10_+_105787935 | 0.13 |
ENSRNOT00000000266
|
Mettl23
|
methyltransferase like 23 |
| chr8_-_12993651 | 0.13 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr8_-_55037604 | 0.13 |
ENSRNOT00000059169
|
Sdhd
|
succinate dehydrogenase complex subunit D |
| chr17_-_21705773 | 0.13 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr17_+_43734461 | 0.13 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr10_+_34975708 | 0.13 |
ENSRNOT00000064984
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr10_+_83856280 | 0.13 |
ENSRNOT00000008964
|
Snf8
|
SNF8, ESCRT-II complex subunit |
| chr2_-_164684985 | 0.13 |
ENSRNOT00000057504
|
Rarres1
|
retinoic acid receptor responder 1 |
| chr7_-_3246071 | 0.13 |
ENSRNOT00000044292
|
Ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr1_+_276309927 | 0.13 |
ENSRNOT00000067460
ENSRNOT00000066236 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr7_-_73450262 | 0.13 |
ENSRNOT00000006943
|
Nipal2
|
NIPA-like domain containing 2 |
| chr7_+_11784341 | 0.13 |
ENSRNOT00000026155
|
Plekhj1
|
pleckstrin homology domain containing J1 |
| chr15_-_83494423 | 0.13 |
ENSRNOT00000037588
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr15_+_45422010 | 0.12 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chrX_+_115561332 | 0.12 |
ENSRNOT00000076680
ENSRNOT00000075912 ENSRNOT00000007968 |
Alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr10_-_3321404 | 0.12 |
ENSRNOT00000093722
ENSRNOT00000003284 |
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr19_+_26194465 | 0.12 |
ENSRNOT00000087627
ENSRNOT00000065931 |
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr8_-_72204730 | 0.12 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr3_+_94848823 | 0.12 |
ENSRNOT00000041362
|
Ccdc73
|
coiled-coil domain containing 73 |
| chr1_-_263920934 | 0.12 |
ENSRNOT00000090755
ENSRNOT00000017006 |
Bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr5_-_154394023 | 0.12 |
ENSRNOT00000079823
|
Rpl11
|
ribosomal protein L11 |
| chr20_+_26755911 | 0.12 |
ENSRNOT00000083017
|
Herc4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr3_-_168033457 | 0.12 |
ENSRNOT00000055111
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr3_-_148010176 | 0.12 |
ENSRNOT00000031683
|
Defb29
|
defensin beta 29 |
| chr19_-_37281933 | 0.12 |
ENSRNOT00000059663
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr2_+_98252925 | 0.12 |
ENSRNOT00000011579
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr3_+_72080630 | 0.12 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr19_+_39357791 | 0.12 |
ENSRNOT00000086435
ENSRNOT00000015006 |
Cyb5b
|
cytochrome b5 type B |
| chr5_-_57267002 | 0.12 |
ENSRNOT00000011455
|
Bag1
|
Bcl2 associated athanogene 1 |
| chr2_+_60131776 | 0.12 |
ENSRNOT00000080786
|
Prlr
|
prolactin receptor |
| chr7_-_145062956 | 0.12 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr1_+_197999037 | 0.12 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr1_+_80056755 | 0.12 |
ENSRNOT00000021221
|
Snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr3_-_111037166 | 0.12 |
ENSRNOT00000017070
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr14_+_115391609 | 0.12 |
ENSRNOT00000079169
|
Asb3
|
ankyrin repeat and SOCS box-containing 3 |
| chr6_+_129538982 | 0.12 |
ENSRNOT00000083626
ENSRNOT00000082522 |
Ak7
|
adenylate kinase 7 |
| chr1_-_222167447 | 0.12 |
ENSRNOT00000028687
|
Prdx5
|
peroxiredoxin 5 |
| chr5_+_2042991 | 0.12 |
ENSRNOT00000050236
|
Eloc
|
elongin C |
| chr5_+_147803735 | 0.12 |
ENSRNOT00000074235
|
Tmem234
|
transmembrane protein 234 |
| chr6_+_127333590 | 0.12 |
ENSRNOT00000063970
|
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr5_+_126783061 | 0.12 |
ENSRNOT00000013224
|
Tmem59
|
transmembrane protein 59 |
| chr4_-_81968832 | 0.12 |
ENSRNOT00000016608
|
Skap2
|
src kinase associated phosphoprotein 2 |
| chr11_+_1896209 | 0.12 |
ENSRNOT00000000909
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr14_-_114047527 | 0.12 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr12_-_11144754 | 0.12 |
ENSRNOT00000084637
|
Zfp655
|
zinc finger protein 655 |
| chr5_+_122847645 | 0.12 |
ENSRNOT00000009516
|
Oma1
|
OMA1 zinc metallopeptidase |
| chr7_-_121783435 | 0.12 |
ENSRNOT00000034912
|
Enthd1
|
ENTH domain containing 1 |
| chr3_+_141225290 | 0.11 |
ENSRNOT00000016351
|
Xrn2
|
5'-3' exoribonuclease 2 |
| chr8_+_70994563 | 0.11 |
ENSRNOT00000051504
ENSRNOT00000077163 |
Spg21
|
spastic paraplegia 21 homolog (human) |
| chr11_-_14160697 | 0.11 |
ENSRNOT00000081096
|
Hspa13
|
heat shock protein 70 family, member 13 |
| chr1_+_56949712 | 0.11 |
ENSRNOT00000020889
|
LOC100125364
|
hypothetical protein LOC100125364 |
| chr1_+_86972244 | 0.11 |
ENSRNOT00000036907
|
Rinl
|
Ras and Rab interactor-like |
| chr3_-_82756953 | 0.11 |
ENSRNOT00000044121
|
Accs
|
1-aminocyclopropane-1-carboxylate synthase homolog |
| chr16_+_21275311 | 0.11 |
ENSRNOT00000027980
|
LOC100911483
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-like |
| chr6_+_2969333 | 0.11 |
ENSRNOT00000047356
|
Morn2
|
MORN repeat containing 2 |
| chr8_+_119137728 | 0.11 |
ENSRNOT00000078170
|
Prss50
|
protease, serine, 50 |
| chr20_+_3344870 | 0.11 |
ENSRNOT00000001058
ENSRNOT00000087613 ENSRNOT00000089978 |
Mrps18b
|
mitochondrial ribosomal protein S18B |
| chr1_-_199823386 | 0.11 |
ENSRNOT00000027375
|
Rgs10
|
regulator of G-protein signaling 10 |
| chr20_-_7654428 | 0.11 |
ENSRNOT00000045044
|
Tcp11
|
t-complex 11 |
| chr18_-_16009710 | 0.11 |
ENSRNOT00000022169
|
Ino80c
|
INO80 complex subunit C |
| chr10_+_39875371 | 0.11 |
ENSRNOT00000013481
|
Rapgef6
|
Rap guanine nucleotide exchange factor 6 |
| chr6_+_33786627 | 0.11 |
ENSRNOT00000008359
|
Pum2
|
pumilio RNA-binding family member 2 |
| chr2_+_189400696 | 0.11 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr2_-_23260965 | 0.11 |
ENSRNOT00000046772
|
LOC103691423
|
60S ribosomal protein L36 pseudogene |
| chr2_+_148722231 | 0.11 |
ENSRNOT00000018022
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr10_+_95242986 | 0.11 |
ENSRNOT00000065589
ENSRNOT00000080460 |
RGD1565033
|
similar to hypothetical protein LOC284018 isoform b |
| chr6_+_10594122 | 0.11 |
ENSRNOT00000020534
|
Cript
|
CXXC repeat containing interactor of PDZ3 domain |
| chr3_+_148327965 | 0.11 |
ENSRNOT00000010851
ENSRNOT00000088481 |
Tpx2
|
TPX2, microtubule nucleation factor |
| chr2_+_86971147 | 0.11 |
ENSRNOT00000032909
|
Zfp458
|
zinc finger protein 458 |
| chr13_-_78852160 | 0.11 |
ENSRNOT00000039082
ENSRNOT00000076514 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr3_+_138597638 | 0.11 |
ENSRNOT00000031623
|
Zfp133
|
zinc finger protein 133 |
| chr1_+_227592635 | 0.11 |
ENSRNOT00000072135
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr5_+_147535525 | 0.11 |
ENSRNOT00000074628
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr1_+_142050458 | 0.11 |
ENSRNOT00000018107
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr3_-_56030744 | 0.11 |
ENSRNOT00000089637
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr1_-_142720257 | 0.11 |
ENSRNOT00000032294
|
Wdr73
|
WD repeat domain 73 |
| chr3_+_123209608 | 0.11 |
ENSRNOT00000028838
|
Itpa
|
inosine triphosphatase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Elf5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.2 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.4 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.1 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 0.1 | 0.2 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 0.1 | 0.2 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.1 | 0.2 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.2 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 0.1 | 0.2 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.1 | GO:2000078 | positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.0 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.0 | 0.1 | GO:0043132 | NAD transport(GO:0043132) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.0 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.2 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0019661 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:1904638 | response to resveratrol(GO:1904638) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.0 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0006296 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0009202 | dTTP biosynthetic process(GO:0006235) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.0 | 0.0 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.0 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.4 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.0 | 0.0 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.0 | 0.0 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:1990910 | response to hypobaric hypoxia(GO:1990910) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.0 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.0 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.0 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 2.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.1 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |