Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
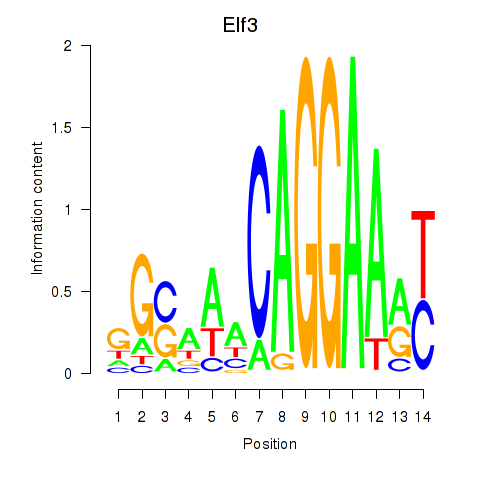
Results for Elf3
Z-value: 0.42
Transcription factors associated with Elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Elf3
|
ENSRNOG00000006330 | E74-like factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elf3 | rn6_v1_chr13_-_52088780_52088780 | -0.67 | 2.2e-01 | Click! |
Activity profile of Elf3 motif
Sorted Z-values of Elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_19791832 | 0.19 |
ENSRNOT00000040393
|
Ushbp1
|
USH1 protein network component harmonin binding protein 1 |
| chr10_-_64642292 | 0.17 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr10_+_47930633 | 0.15 |
ENSRNOT00000003515
|
Grap
|
GRB2-related adaptor protein |
| chr5_-_169630340 | 0.14 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr3_+_177226417 | 0.13 |
ENSRNOT00000031328
|
Oprl1
|
opioid related nociceptin receptor 1 |
| chr4_-_78342863 | 0.13 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr7_-_93826665 | 0.13 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr20_-_27301952 | 0.13 |
ENSRNOT00000000438
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr14_+_86652365 | 0.12 |
ENSRNOT00000077428
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr8_-_128754514 | 0.12 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr1_-_43638161 | 0.11 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_-_59688692 | 0.10 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr12_+_38144855 | 0.10 |
ENSRNOT00000032274
|
Hcar1
|
hydroxycarboxylic acid receptor 1 |
| chr4_+_51614676 | 0.09 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr9_-_81864202 | 0.09 |
ENSRNOT00000084964
|
Zfp142
|
zinc finger protein 142 |
| chr10_+_40553180 | 0.09 |
ENSRNOT00000087763
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr14_+_109533792 | 0.09 |
ENSRNOT00000067690
|
LOC108348154
|
ankyrin repeat domain-containing protein 29-like |
| chr13_+_105684420 | 0.09 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr19_+_25043680 | 0.09 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr3_-_148932878 | 0.08 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr2_+_45668969 | 0.08 |
ENSRNOT00000014761
ENSRNOT00000071353 |
Arl15
|
ADP-ribosylation factor like GTPase 15 |
| chr10_-_16046033 | 0.08 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_-_173653905 | 0.07 |
ENSRNOT00000038556
|
Plekhn1
|
pleckstrin homology domain containing N1 |
| chr2_-_204341676 | 0.07 |
ENSRNOT00000021713
|
Slc22a15
|
solute carrier family 22, member 15 |
| chr6_+_12415805 | 0.07 |
ENSRNOT00000022380
|
Gtf2a1l
|
general transcription factor 2A subunit 1 like |
| chr10_-_85636902 | 0.07 |
ENSRNOT00000017241
|
Pcgf2
|
polycomb group ring finger 2 |
| chr8_+_48925604 | 0.07 |
ENSRNOT00000077445
|
Ddx6
|
DEAD-box helicase 6 |
| chr1_-_205040755 | 0.07 |
ENSRNOT00000023574
|
Ctbp2
|
C-terminal binding protein 2 |
| chr2_+_207262934 | 0.07 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr18_-_30775401 | 0.07 |
ENSRNOT00000027128
|
Slc25a2
|
solute carrier family 25 member 2 |
| chr3_+_107142762 | 0.07 |
ENSRNOT00000006177
|
RGD1563680
|
similar to CDNA sequence BC052040 |
| chr1_-_215846911 | 0.07 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr14_+_33624390 | 0.07 |
ENSRNOT00000002907
|
Aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr20_+_27954433 | 0.07 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr7_-_66909470 | 0.06 |
ENSRNOT00000066381
|
Ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr7_-_117680004 | 0.06 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr5_-_158439078 | 0.06 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr1_-_4011161 | 0.06 |
ENSRNOT00000044778
|
Stxbp5
|
syntaxin binding protein 5 |
| chr14_+_84150908 | 0.06 |
ENSRNOT00000005516
|
Dusp18
|
dual specificity phosphatase 18 |
| chr11_+_57430166 | 0.06 |
ENSRNOT00000093201
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chrX_+_22247088 | 0.06 |
ENSRNOT00000079802
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr19_-_58735173 | 0.06 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr10_+_86337728 | 0.06 |
ENSRNOT00000085408
|
Tcap
|
titin-cap |
| chr9_-_45206427 | 0.06 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr7_-_98098268 | 0.06 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr10_+_55927223 | 0.06 |
ENSRNOT00000011523
|
Kcnab3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3 |
| chr1_-_198316882 | 0.06 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr20_+_3363737 | 0.06 |
ENSRNOT00000079654
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr16_+_78539489 | 0.06 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr10_+_17421075 | 0.05 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr3_-_162579201 | 0.05 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr10_+_73868943 | 0.05 |
ENSRNOT00000081012
|
Tubd1
|
tubulin, delta 1 |
| chr1_-_4011511 | 0.05 |
ENSRNOT00000040559
|
Stxbp5
|
syntaxin binding protein 5 |
| chrX_+_134979646 | 0.05 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr17_-_18592750 | 0.05 |
ENSRNOT00000065742
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr14_+_38192446 | 0.05 |
ENSRNOT00000090242
|
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr4_-_51586881 | 0.05 |
ENSRNOT00000064983
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr7_-_130151414 | 0.05 |
ENSRNOT00000010467
|
Plxnb2
|
plexin B2 |
| chr16_+_71058022 | 0.05 |
ENSRNOT00000066901
|
Bag4
|
BCL2-associated athanogene 4 |
| chr1_+_211351350 | 0.05 |
ENSRNOT00000082751
ENSRNOT00000030461 |
Jakmip3
|
janus kinase and microtubule interacting protein 3 |
| chr13_+_99136871 | 0.05 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr20_+_9948908 | 0.05 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr3_+_163570532 | 0.05 |
ENSRNOT00000010054
|
Arfgef2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr15_-_42898150 | 0.05 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr15_-_33218456 | 0.05 |
ENSRNOT00000017753
|
Ajuba
|
ajuba LIM protein |
| chr3_+_113918629 | 0.05 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr14_-_5006594 | 0.05 |
ENSRNOT00000076571
|
Zfp326
|
zinc finger protein 326 |
| chr7_-_122329443 | 0.04 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr17_-_27602934 | 0.04 |
ENSRNOT00000079298
|
Rreb1
|
ras responsive element binding protein 1 |
| chr1_-_175586802 | 0.04 |
ENSRNOT00000071333
|
AABR07005027.1
|
|
| chr1_-_261179790 | 0.04 |
ENSRNOT00000074420
ENSRNOT00000072073 |
Exosc1
|
exosome component 1 |
| chr2_+_12516702 | 0.04 |
ENSRNOT00000050072
|
Tmem161b
|
transmembrane protein 161B |
| chr2_-_53300404 | 0.04 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr1_-_89509343 | 0.04 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr3_-_112174269 | 0.04 |
ENSRNOT00000067836
|
Tmem87a
|
transmembrane protein 87A |
| chrX_-_1952111 | 0.04 |
ENSRNOT00000061140
|
Jade3
|
jade family PHD finger 3 |
| chr1_-_189549891 | 0.04 |
ENSRNOT00000019747
|
Eri2
|
ERI1 exoribonuclease family member 2 |
| chr10_+_104437648 | 0.04 |
ENSRNOT00000035001
|
AC130970.1
|
|
| chr19_+_9668186 | 0.04 |
ENSRNOT00000016563
|
Cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr4_-_183544850 | 0.04 |
ENSRNOT00000071407
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr4_-_163214678 | 0.04 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr3_-_63568464 | 0.04 |
ENSRNOT00000068494
|
AABR07052585.2
|
|
| chr18_+_30592794 | 0.04 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr9_-_110936631 | 0.04 |
ENSRNOT00000064431
|
Fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chr1_-_219682287 | 0.04 |
ENSRNOT00000026092
|
Rhod
|
ras homolog family member D |
| chr2_-_183582553 | 0.04 |
ENSRNOT00000014236
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr19_+_30668602 | 0.04 |
ENSRNOT00000035084
|
Usp38
|
ubiquitin specific peptidase 38 |
| chr19_-_37528011 | 0.04 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr1_+_215605404 | 0.04 |
ENSRNOT00000027454
|
Syt8
|
synaptotagmin 8 |
| chr15_-_56970365 | 0.04 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr4_+_163162211 | 0.04 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr4_+_167754525 | 0.04 |
ENSRNOT00000007889
|
Etv6
|
ets variant 6 |
| chr7_+_70753101 | 0.04 |
ENSRNOT00000090001
|
R3hdm2
|
R3H domain containing 2 |
| chr5_-_159119171 | 0.04 |
ENSRNOT00000074074
ENSRNOT00000075382 ENSRNOT00000074378 |
Arhgef10l
|
Rho guanine nucleotide exchange factor 10 like |
| chrX_-_115175299 | 0.04 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr4_+_78310379 | 0.04 |
ENSRNOT00000032802
|
Gimap9
|
GTPase, IMAP family member 9 |
| chr11_+_84745904 | 0.03 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr9_+_121802673 | 0.03 |
ENSRNOT00000086534
|
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr18_-_399242 | 0.03 |
ENSRNOT00000045926
|
F8
|
coagulation factor VIII |
| chr8_-_84632817 | 0.03 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr4_+_78371121 | 0.03 |
ENSRNOT00000059157
|
Gimap1
|
GTPase, IMAP family member 1 |
| chr10_-_12916784 | 0.03 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr7_-_107616038 | 0.03 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr15_+_4240203 | 0.03 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr4_+_71652354 | 0.03 |
ENSRNOT00000022672
ENSRNOT00000022543 |
Casp2
|
caspase 2 |
| chr17_+_5225835 | 0.03 |
ENSRNOT00000022373
|
Zcchc6
|
zinc finger CCHC-type containing 6 |
| chr7_-_60825242 | 0.03 |
ENSRNOT00000009203
|
Nup107
|
nucleoporin 107 |
| chr10_-_85084850 | 0.03 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr10_+_45322248 | 0.03 |
ENSRNOT00000047268
|
Trim11
|
tripartite motif-containing 11 |
| chr8_+_33239139 | 0.03 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr17_-_75886523 | 0.03 |
ENSRNOT00000046266
|
Usp6nl
|
USP6 N-terminal like |
| chr17_+_11683862 | 0.03 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr7_+_130326600 | 0.03 |
ENSRNOT00000084776
ENSRNOT00000055808 |
Ncaph2
|
non-SMC condensin II complex, subunit H2 |
| chr8_+_42443442 | 0.03 |
ENSRNOT00000015986
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr13_-_37287458 | 0.03 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr7_+_122818975 | 0.03 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chr3_-_111266542 | 0.03 |
ENSRNOT00000031672
|
Ino80
|
INO80 complex subunit |
| chr6_+_3012804 | 0.03 |
ENSRNOT00000061980
|
Arhgef33
|
Rho guanine nucleotide exchange factor 33 |
| chr10_+_39109522 | 0.03 |
ENSRNOT00000010968
|
Irf1
|
interferon regulatory factor 1 |
| chr17_-_46794845 | 0.03 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr3_-_92242318 | 0.03 |
ENSRNOT00000007018
|
Trim44
|
tripartite motif-containing 44 |
| chr2_+_18937458 | 0.03 |
ENSRNOT00000022377
|
Tmem167a
|
transmembrane protein 167A |
| chr14_-_18076907 | 0.03 |
ENSRNOT00000003472
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr10_-_98018014 | 0.03 |
ENSRNOT00000005367
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr5_+_60250546 | 0.03 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr13_+_88606894 | 0.03 |
ENSRNOT00000048692
|
Sh2d1b
|
SH2 domain containing 1B |
| chr13_-_78957183 | 0.03 |
ENSRNOT00000076954
ENSRNOT00000003851 |
Klhl20
|
kelch-like family member 20 |
| chr17_-_63145577 | 0.03 |
ENSRNOT00000074389
|
Gtpbp4
|
GTP binding protein 4 |
| chr10_+_85608544 | 0.03 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr8_+_49378644 | 0.03 |
ENSRNOT00000007588
|
Jaml
|
junction adhesion molecule like |
| chr3_-_147487170 | 0.03 |
ENSRNOT00000077580
|
Fam110a
|
family with sequence similarity 110, member A |
| chr9_-_44560837 | 0.03 |
ENSRNOT00000090870
|
Mitd1
|
microtubule interacting and trafficking domain containing 1 |
| chr15_-_52288893 | 0.03 |
ENSRNOT00000016209
|
Fam160b2
|
family with sequence similarity 160, member B2 |
| chr10_-_65437143 | 0.03 |
ENSRNOT00000017264
|
Nek8
|
NIMA-related kinase 8 |
| chr18_-_28017925 | 0.03 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr8_-_71533069 | 0.03 |
ENSRNOT00000021863
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr19_-_10360310 | 0.03 |
ENSRNOT00000087100
|
Katnb1
|
katanin regulatory subunit B1 |
| chrX_+_65566047 | 0.03 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr1_-_85368031 | 0.03 |
ENSRNOT00000026778
|
Paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr12_+_1889331 | 0.02 |
ENSRNOT00000066281
|
Arhgef18
|
Rho/Rac guanine nucleotide exchange factor 18 |
| chr12_-_31249118 | 0.02 |
ENSRNOT00000058586
ENSRNOT00000073105 |
Adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr1_+_64046377 | 0.02 |
ENSRNOT00000085010
|
Tmc4
|
transmembrane channel-like 4 |
| chrX_-_116792707 | 0.02 |
ENSRNOT00000049482
|
Amot
|
angiomotin |
| chrX_+_105500173 | 0.02 |
ENSRNOT00000040476
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr13_-_82753438 | 0.02 |
ENSRNOT00000075948
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr14_+_83343330 | 0.02 |
ENSRNOT00000089198
|
Pisd
|
phosphatidylserine decarboxylase |
| chr14_+_5928737 | 0.02 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr3_-_112125290 | 0.02 |
ENSRNOT00000011155
ENSRNOT00000085132 |
Vps39
|
VPS39 HOPS complex subunit |
| chr7_+_117963740 | 0.02 |
ENSRNOT00000075405
|
LOC108348189
|
COMM domain-containing protein 5 |
| chr12_+_11179329 | 0.02 |
ENSRNOT00000001302
|
Zfp394
|
zinc finger protein 394 |
| chr9_-_100479868 | 0.02 |
ENSRNOT00000022748
|
Pask
|
PAS domain containing serine/threonine kinase |
| chrX_+_70440059 | 0.02 |
ENSRNOT00000003946
|
Arr3
|
arrestin 3 |
| chr17_-_13586554 | 0.02 |
ENSRNOT00000041371
|
Secisbp2
|
SECIS binding protein 2 |
| chr13_+_52662996 | 0.02 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr10_-_70802782 | 0.02 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr4_-_145948996 | 0.02 |
ENSRNOT00000043476
|
Atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr16_+_53957858 | 0.02 |
ENSRNOT00000013078
|
Frg1
|
FSHD region gene 1 |
| chr7_-_30397930 | 0.02 |
ENSRNOT00000010414
|
Actr6
|
ARP6 actin-related protein 6 homolog |
| chr4_+_62380914 | 0.02 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chr14_-_86796378 | 0.02 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr8_+_12823155 | 0.02 |
ENSRNOT00000011008
|
Sesn3
|
sestrin 3 |
| chr10_+_110346453 | 0.02 |
ENSRNOT00000054928
|
Tex19.1
|
testis expressed 19.1 |
| chr2_-_185303610 | 0.02 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr12_-_48663143 | 0.02 |
ENSRNOT00000082809
|
Ficd
|
FIC domain containing |
| chr2_+_127625683 | 0.02 |
ENSRNOT00000015474
|
Hspa4l
|
heat shock protein 4-like |
| chr4_-_118595580 | 0.02 |
ENSRNOT00000024436
|
Anxa4
|
annexin A4 |
| chr11_-_4332255 | 0.02 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr7_+_70987861 | 0.02 |
ENSRNOT00000072906
|
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr1_+_63842277 | 0.02 |
ENSRNOT00000087957
ENSRNOT00000080820 |
Lilrb3a
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3A |
| chr1_+_261180007 | 0.02 |
ENSRNOT00000072181
|
Zdhhc16
|
zinc finger, DHHC-type containing 16 |
| chr7_-_60309878 | 0.02 |
ENSRNOT00000007610
|
RGD1306474
|
similar to RIKEN cDNA 9530003J23 |
| chr4_-_170092848 | 0.02 |
ENSRNOT00000072147
|
Hist2h4
|
histone cluster 2, H4 |
| chr16_-_10661528 | 0.02 |
ENSRNOT00000085012
|
Fam35a
|
family with sequence similarity 35, member A |
| chr3_-_15278645 | 0.02 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr1_-_157461588 | 0.02 |
ENSRNOT00000068402
|
Ankrd42
|
ankyrin repeat domain 42 |
| chr3_-_48535909 | 0.02 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr6_+_94636222 | 0.02 |
ENSRNOT00000005846
|
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr6_+_109939345 | 0.02 |
ENSRNOT00000013560
|
Ift43
|
intraflagellar transport 43 |
| chr10_-_34166599 | 0.02 |
ENSRNOT00000003246
|
Trim41
|
tripartite motif-containing 41 |
| chr7_+_117773948 | 0.02 |
ENSRNOT00000021673
|
Lrrc14
|
leucine rich repeat containing 14 |
| chr4_-_77747070 | 0.02 |
ENSRNOT00000009247
|
Zfp746
|
zinc finger protein 746 |
| chr5_-_2882668 | 0.02 |
ENSRNOT00000049741
|
Terf1
|
telomeric repeat binding factor 1 |
| chr12_+_23514981 | 0.02 |
ENSRNOT00000001937
|
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chrX_+_24891156 | 0.01 |
ENSRNOT00000071173
|
Wwc3
|
WWC family member 3 |
| chr15_+_18399733 | 0.01 |
ENSRNOT00000061158
|
Fam107a
|
family with sequence similarity 107, member A |
| chr15_+_18399515 | 0.01 |
ENSRNOT00000071273
|
Fam107a
|
family with sequence similarity 107, member A |
| chr1_+_189550354 | 0.01 |
ENSRNOT00000083153
|
Exnef
|
exonuclease NEF-sp |
| chr15_+_34270648 | 0.01 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr15_-_34469350 | 0.01 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr12_-_45182974 | 0.01 |
ENSRNOT00000001503
|
Taok3
|
TAO kinase 3 |
| chr5_+_120498883 | 0.01 |
ENSRNOT00000007859
|
Leprot
|
leptin receptor overlapping transcript |
| chr1_-_191007503 | 0.01 |
ENSRNOT00000023262
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr7_-_60341264 | 0.01 |
ENSRNOT00000007747
|
Lyz2
|
lysozyme 2 |
| chr15_+_87704340 | 0.01 |
ENSRNOT00000034987
|
Scel
|
sciellin |
| chr1_+_221735517 | 0.01 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr3_+_138974871 | 0.01 |
ENSRNOT00000012524
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr18_-_786674 | 0.01 |
ENSRNOT00000021955
|
Cetn1
|
centrin 1 |
| chr5_+_172986291 | 0.01 |
ENSRNOT00000022900
|
Nadk
|
NAD kinase |
| chr13_+_99005142 | 0.01 |
ENSRNOT00000004293
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr10_+_12046541 | 0.01 |
ENSRNOT00000081191
|
Mefv
|
MEFV, pyrin innate immunity regulator |
Network of associatons between targets according to the STRING database.
First level regulatory network of Elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.0 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.0 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0004903 | growth hormone receptor activity(GO:0004903) |