Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
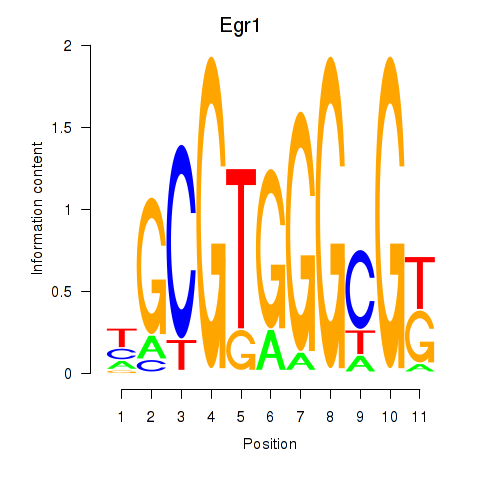
Results for Egr1
Z-value: 0.94
Transcription factors associated with Egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr1
|
ENSRNOG00000019422 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | rn6_v1_chr18_+_27657628_27657628 | 0.81 | 9.7e-02 | Click! |
Activity profile of Egr1 motif
Sorted Z-values of Egr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_115910522 | 0.58 |
ENSRNOT00000076998
ENSRNOT00000067442 |
Arc
|
activity-regulated cytoskeleton-associated protein |
| chr6_+_135856218 | 0.53 |
ENSRNOT00000066715
|
RGD1560608
|
similar to novel protein |
| chr8_-_64572850 | 0.36 |
ENSRNOT00000015415
|
Senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr5_+_58393603 | 0.36 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr10_+_109107389 | 0.35 |
ENSRNOT00000068437
ENSRNOT00000005687 |
Baiap2
|
BAI1-associated protein 2 |
| chr5_+_58393233 | 0.33 |
ENSRNOT00000000142
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr8_+_96551245 | 0.31 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr3_+_170550314 | 0.31 |
ENSRNOT00000006991
|
Tfap2c
|
transcription factor AP-2 gamma |
| chrX_+_13117239 | 0.29 |
ENSRNOT00000088998
|
AABR07037112.1
|
|
| chr7_+_142877086 | 0.28 |
ENSRNOT00000088730
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chr9_+_53906073 | 0.28 |
ENSRNOT00000017813
|
Nab1
|
Ngfi-A binding protein 1 |
| chr1_+_101517714 | 0.26 |
ENSRNOT00000028423
|
Plekha4
|
pleckstrin homology domain containing A4 |
| chr7_+_122456799 | 0.23 |
ENSRNOT00000025564
|
Mchr1
|
melanin-concentrating hormone receptor 1 |
| chr8_-_128266639 | 0.23 |
ENSRNOT00000064555
ENSRNOT00000066932 |
Scn5a
|
sodium voltage-gated channel alpha subunit 5 |
| chr8_-_40023193 | 0.23 |
ENSRNOT00000014404
|
Nrgn
|
neurogranin |
| chr2_+_113984824 | 0.22 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr9_-_104350308 | 0.21 |
ENSRNOT00000033958
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr2_-_188645196 | 0.21 |
ENSRNOT00000083793
|
Efna3
|
ephrin A3 |
| chr4_-_16130563 | 0.21 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr16_-_9709347 | 0.21 |
ENSRNOT00000083933
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr4_+_25825567 | 0.20 |
ENSRNOT00000009417
|
Cdk14
|
cyclin-dependent kinase 14 |
| chr2_+_113984646 | 0.20 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr20_+_4572100 | 0.20 |
ENSRNOT00000000476
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr3_-_110492398 | 0.19 |
ENSRNOT00000056450
|
Ankrd63
|
ankyrin repeat domain 63 |
| chrX_+_1321315 | 0.19 |
ENSRNOT00000014250
|
Syn1
|
synapsin I |
| chr10_-_88667345 | 0.19 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr1_-_222590112 | 0.19 |
ENSRNOT00000028763
ENSRNOT00000091990 |
Mark2
|
microtubule affinity regulating kinase 2 |
| chr7_-_84023316 | 0.18 |
ENSRNOT00000005556
|
Kcnv1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr5_-_36683356 | 0.18 |
ENSRNOT00000009043
|
Pou3f2
|
POU class 3 homeobox 2 |
| chr8_-_120446455 | 0.18 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr10_-_37645802 | 0.18 |
ENSRNOT00000008022
|
Tcf7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr9_+_54212767 | 0.17 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr11_+_86512797 | 0.17 |
ENSRNOT00000051680
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr12_-_22680630 | 0.16 |
ENSRNOT00000041808
|
Vgf
|
VGF nerve growth factor inducible |
| chr7_+_12006710 | 0.16 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr1_-_141349881 | 0.16 |
ENSRNOT00000021109
|
Rhcg
|
Rh family, C glycoprotein |
| chr4_-_16130848 | 0.16 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr16_-_6404578 | 0.16 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_-_186245771 | 0.16 |
ENSRNOT00000066004
ENSRNOT00000079954 |
Dclk2
|
doublecortin-like kinase 2 |
| chrX_-_38196060 | 0.15 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr7_-_140437467 | 0.14 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr10_+_85744568 | 0.14 |
ENSRNOT00000005522
|
Lasp1
|
LIM and SH3 protein 1 |
| chr19_-_11341863 | 0.14 |
ENSRNOT00000025694
|
Mt4
|
metallothionein 4 |
| chr11_+_69484293 | 0.14 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr17_+_9653561 | 0.14 |
ENSRNOT00000018899
|
Pdlim7
|
PDZ and LIM domain 7 |
| chr11_+_83868655 | 0.14 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr11_-_83546674 | 0.14 |
ENSRNOT00000044896
|
Ephb3
|
Eph receptor B3 |
| chr16_-_6405117 | 0.14 |
ENSRNOT00000047737
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_-_186245342 | 0.14 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr3_-_111560556 | 0.14 |
ENSRNOT00000030532
|
Ltk
|
leukocyte receptor tyrosine kinase |
| chr7_+_41115154 | 0.13 |
ENSRNOT00000066174
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr6_+_10912383 | 0.13 |
ENSRNOT00000061747
ENSRNOT00000086247 |
Ttc7a
|
tetratricopeptide repeat domain 7A |
| chr10_-_13515448 | 0.13 |
ENSRNOT00000067660
|
Pdpk1
|
3-phosphoinositide dependent protein kinase-1 |
| chr7_-_36499784 | 0.13 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr19_-_55490426 | 0.13 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr8_-_97115027 | 0.13 |
ENSRNOT00000018685
|
Ankrd34c
|
ankyrin repeat domain 34C |
| chr1_-_78440738 | 0.13 |
ENSRNOT00000020855
|
Npas1
|
neuronal PAS domain protein 1 |
| chr3_-_123376455 | 0.12 |
ENSRNOT00000028842
|
RGD1565616
|
RGD1565616 |
| chr11_+_82466071 | 0.12 |
ENSRNOT00000036958
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr16_-_6404957 | 0.12 |
ENSRNOT00000048459
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr7_-_121232741 | 0.12 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr8_+_22050222 | 0.12 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr1_+_220428481 | 0.11 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chrX_+_122808605 | 0.11 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr1_-_101360971 | 0.11 |
ENSRNOT00000028164
|
Lin7b
|
lin-7 homolog B, crumbs cell polarity complex component |
| chr7_+_73588163 | 0.11 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr9_+_71230108 | 0.11 |
ENSRNOT00000018326
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr17_+_9746485 | 0.11 |
ENSRNOT00000072298
|
Pfn3
|
profilin 3 |
| chr1_-_103256823 | 0.11 |
ENSRNOT00000018860
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr2_+_198721724 | 0.11 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr1_-_64350338 | 0.11 |
ENSRNOT00000078444
|
Cacng8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr6_-_29483460 | 0.11 |
ENSRNOT00000007160
|
Klhl29
|
kelch-like family member 29 |
| chr2_-_34185682 | 0.11 |
ENSRNOT00000066925
ENSRNOT00000082755 |
Nln
|
neurolysin |
| chrX_-_15467875 | 0.11 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr1_-_7064870 | 0.11 |
ENSRNOT00000019983
|
Stx11
|
syntaxin 11 |
| chrX_+_39711201 | 0.11 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr18_-_31071371 | 0.11 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chr16_+_71629525 | 0.11 |
ENSRNOT00000035347
ENSRNOT00000088462 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr10_+_55687050 | 0.11 |
ENSRNOT00000057136
|
Per1
|
period circadian clock 1 |
| chr6_+_132246602 | 0.10 |
ENSRNOT00000009896
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr7_-_59514939 | 0.10 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chrX_-_30831483 | 0.10 |
ENSRNOT00000004443
|
Glra2
|
glycine receptor, alpha 2 |
| chr1_-_80544825 | 0.10 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr11_+_61661724 | 0.10 |
ENSRNOT00000079423
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr8_+_22189600 | 0.10 |
ENSRNOT00000061100
|
Pde4a
|
phosphodiesterase 4A |
| chrX_+_120859968 | 0.10 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr2_+_34186091 | 0.10 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr10_-_85517683 | 0.10 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr4_-_90882285 | 0.10 |
ENSRNOT00000039247
|
Snca
|
synuclein alpha |
| chr10_+_56627411 | 0.10 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr4_+_117215064 | 0.10 |
ENSRNOT00000020909
|
Smyd5
|
SMYD family member 5 |
| chr12_+_22665112 | 0.09 |
ENSRNOT00000001918
|
Ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr7_+_140464999 | 0.09 |
ENSRNOT00000090156
|
Wnt1
|
wingless-type MMTV integration site family, member 1 |
| chr20_+_4576057 | 0.09 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr17_+_82065937 | 0.09 |
ENSRNOT00000051349
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr1_-_174495258 | 0.09 |
ENSRNOT00000052377
ENSRNOT00000086349 |
Scube2
|
signal peptide, CUB domain and EGF like domain containing 2 |
| chr7_+_120125633 | 0.09 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr3_+_2396143 | 0.09 |
ENSRNOT00000012388
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr20_-_4879779 | 0.09 |
ENSRNOT00000081924
|
Hspa1b
|
heat shock protein family A (Hsp70) member 1B |
| chr7_-_105592804 | 0.09 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr3_-_163935617 | 0.09 |
ENSRNOT00000074023
|
Kcnb1
|
potassium voltage-gated channel subfamily B member 1 |
| chr1_+_220335254 | 0.09 |
ENSRNOT00000072261
|
Rin1
|
Ras and Rab interactor 1 |
| chr5_+_148051126 | 0.09 |
ENSRNOT00000074415
|
Ptp4a2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr20_-_2701637 | 0.09 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr11_+_61662270 | 0.09 |
ENSRNOT00000079521
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr7_-_14302552 | 0.09 |
ENSRNOT00000091368
|
Brd4
|
bromodomain containing 4 |
| chr1_+_82480195 | 0.09 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr6_+_27975849 | 0.09 |
ENSRNOT00000060810
|
Dtnb
|
dystrobrevin, beta |
| chr10_-_103736973 | 0.09 |
ENSRNOT00000004372
|
Nat9
|
N-acetyltransferase 9 |
| chr9_-_119818310 | 0.08 |
ENSRNOT00000019359
|
Smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr20_-_2701815 | 0.08 |
ENSRNOT00000061950
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr14_+_84417320 | 0.08 |
ENSRNOT00000008788
|
Tbc1d10a
|
TBC1 domain family, member 10a |
| chr8_+_128577345 | 0.08 |
ENSRNOT00000082356
|
Wdr48
|
WD repeat domain 48 |
| chr15_-_61772516 | 0.08 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr1_+_165237847 | 0.08 |
ENSRNOT00000022963
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr1_-_82120902 | 0.08 |
ENSRNOT00000027684
|
Erf
|
Ets2 repressor factor |
| chr3_-_92290919 | 0.08 |
ENSRNOT00000007077
|
Fjx1
|
four jointed box 1 |
| chr8_+_53295222 | 0.08 |
ENSRNOT00000009724
ENSRNOT00000067420 |
Usp28
|
ubiquitin specific peptidase 28 |
| chr6_+_27975417 | 0.08 |
ENSRNOT00000077830
|
Dtnb
|
dystrobrevin, beta |
| chr5_-_136112344 | 0.08 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr1_+_91363492 | 0.08 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr16_+_47717951 | 0.08 |
ENSRNOT00000018689
|
Ing2
|
inhibitor of growth family, member 2 |
| chr7_-_70969905 | 0.08 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr9_+_112293388 | 0.08 |
ENSRNOT00000020767
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr8_-_55408464 | 0.08 |
ENSRNOT00000066893
|
Sik2
|
salt-inducible kinase 2 |
| chr12_+_16084266 | 0.08 |
ENSRNOT00000079866
ENSRNOT00000080774 |
Ttyh3
|
tweety family member 3 |
| chr11_-_86890390 | 0.08 |
ENSRNOT00000046183
|
Trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr1_+_144070754 | 0.08 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr12_+_22727335 | 0.08 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr7_-_75422268 | 0.08 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr14_+_15059387 | 0.08 |
ENSRNOT00000002843
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr9_+_47386626 | 0.08 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr10_+_67762995 | 0.07 |
ENSRNOT00000000253
|
Zfp207
|
zinc finger protein 207 |
| chr8_+_118206698 | 0.07 |
ENSRNOT00000082607
ENSRNOT00000064140 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr7_-_14303055 | 0.07 |
ENSRNOT00000008963
|
Brd4
|
bromodomain containing 4 |
| chr9_+_65763437 | 0.07 |
ENSRNOT00000043558
|
Stradb
|
STE20-related kinase adaptor beta |
| chr8_+_117126692 | 0.07 |
ENSRNOT00000083046
ENSRNOT00000085609 |
Usp4
|
ubiquitin specific peptidase 4 |
| chr3_+_108795235 | 0.07 |
ENSRNOT00000007028
|
Spred1
|
sprouty-related, EVH1 domain containing 1 |
| chrX_+_120860178 | 0.07 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr10_+_93520132 | 0.07 |
ENSRNOT00000055137
|
Mrc2
|
mannose receptor, C type 2 |
| chr4_-_153028129 | 0.07 |
ENSRNOT00000074558
|
Cecr6
|
cat eye syndrome chromosome region, candidate 6 |
| chr9_-_44456554 | 0.07 |
ENSRNOT00000080011
|
Tsga10
|
testis specific 10 |
| chr10_-_65766050 | 0.07 |
ENSRNOT00000013639
|
Sarm1
|
sterile alpha and TIR motif containing 1 |
| chr13_+_85110098 | 0.07 |
ENSRNOT00000087368
|
Fam78b
|
family with sequence similarity 78, member B |
| chr1_-_165967069 | 0.07 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr9_+_81783349 | 0.06 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr18_+_70733872 | 0.06 |
ENSRNOT00000067018
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr20_-_3299420 | 0.06 |
ENSRNOT00000090999
|
Gnl1
|
G protein nucleolar 1 |
| chr1_+_190964885 | 0.06 |
ENSRNOT00000039186
|
Mettl9
|
methyltransferase like 9 |
| chr8_+_44990014 | 0.06 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr12_+_22726643 | 0.06 |
ENSRNOT00000040015
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr1_+_64031859 | 0.06 |
ENSRNOT00000090873
|
Mboat7l1
|
membrane bound O-acyltransferase domain containing 7-like 1 |
| chr11_+_86890585 | 0.06 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr20_-_35449280 | 0.06 |
ENSRNOT00000057422
|
Man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr1_+_246954980 | 0.06 |
ENSRNOT00000020272
|
Slc1a1
|
solute carrier family 1 member 1 |
| chr11_-_61499557 | 0.06 |
ENSRNOT00000046208
|
Usf3
|
upstream transcription factor family member 3 |
| chr1_+_264670841 | 0.06 |
ENSRNOT00000034814
ENSRNOT00000082412 |
Slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr3_+_82081906 | 0.06 |
ENSRNOT00000057001
|
Tp53i11
|
tumor protein p53 inducible protein 11 |
| chr7_-_117300662 | 0.06 |
ENSRNOT00000006376
|
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr4_+_122282279 | 0.06 |
ENSRNOT00000038244
|
Zxdc
|
ZXD family zinc finger C |
| chr1_-_72727112 | 0.06 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr1_-_199159125 | 0.06 |
ENSRNOT00000025618
|
Bcl7c
|
BCL tumor suppressor 7C |
| chr9_-_65763273 | 0.06 |
ENSRNOT00000015034
|
Trak2
|
trafficking kinesin protein 2 |
| chr12_-_46920952 | 0.06 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr10_+_55675575 | 0.06 |
ENSRNOT00000057295
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr8_+_64573358 | 0.06 |
ENSRNOT00000083558
|
Myo9a
|
myosin IXA |
| chr6_-_98566523 | 0.06 |
ENSRNOT00000077715
|
Ppp2r5e
|
protein phosphatase 2 regulatory subunit B', epsilon |
| chr1_-_94494980 | 0.06 |
ENSRNOT00000020014
|
Ccne1
|
cyclin E1 |
| chr18_+_29110242 | 0.06 |
ENSRNOT00000025756
|
Pura
|
purine rich element binding protein A |
| chr8_-_48762342 | 0.05 |
ENSRNOT00000049125
|
Foxr1
|
forkhead box R1 |
| chr12_+_22153983 | 0.05 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr3_+_14299479 | 0.05 |
ENSRNOT00000092745
|
Cntrl
|
centriolin |
| chr6_+_109466060 | 0.05 |
ENSRNOT00000011339
|
Jdp2
|
Jun dimerization protein 2 |
| chr7_+_130474279 | 0.05 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr3_-_171134655 | 0.05 |
ENSRNOT00000028960
|
RGD1561252
|
similar to Ubiquitin carboxyl-terminal hydrolase isozyme L3 (Ubiquitin thiolesterase L3) |
| chr3_-_15433252 | 0.05 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
| chr4_-_28953067 | 0.05 |
ENSRNOT00000013989
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chrX_+_157150655 | 0.05 |
ENSRNOT00000090795
|
Pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr11_-_17340373 | 0.05 |
ENSRNOT00000089762
|
AABR07033324.1
|
|
| chr6_+_42568220 | 0.05 |
ENSRNOT00000074829
|
Pdia6
|
protein disulfide isomerase family A, member 6 |
| chrX_-_156379189 | 0.05 |
ENSRNOT00000083148
|
Plxna3
|
plexin A3 |
| chr5_+_57028467 | 0.05 |
ENSRNOT00000010061
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr2_+_209433103 | 0.05 |
ENSRNOT00000024036
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr13_+_51384389 | 0.04 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr8_+_128577080 | 0.04 |
ENSRNOT00000024172
|
Wdr48
|
WD repeat domain 48 |
| chr1_+_116587815 | 0.04 |
ENSRNOT00000021366
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr8_-_48564722 | 0.04 |
ENSRNOT00000067902
|
Cbl
|
Cbl proto-oncogene |
| chr20_-_3299580 | 0.04 |
ENSRNOT00000050373
|
Gnl1
|
G protein nucleolar 1 |
| chr17_+_45467015 | 0.04 |
ENSRNOT00000081408
|
Gpx5
|
glutathione peroxidase 5 |
| chr5_-_99033107 | 0.04 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr3_-_2534663 | 0.04 |
ENSRNOT00000049297
ENSRNOT00000044246 |
Grin1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr11_-_71108184 | 0.04 |
ENSRNOT00000051989
ENSRNOT00000073865 ENSRNOT00000063883 |
Lrch3
|
leucine rich repeats and calponin homology domain containing 3 |
| chr10_-_56531483 | 0.04 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr3_+_177188044 | 0.04 |
ENSRNOT00000022073
|
Tcea2
|
transcription elongation factor A2 |
| chr14_+_83226601 | 0.04 |
ENSRNOT00000072368
|
Prr14l
|
proline rich 14-like |
| chr13_-_96238474 | 0.04 |
ENSRNOT00000044477
ENSRNOT00000078061 |
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U |
| chr12_-_22726982 | 0.04 |
ENSRNOT00000001921
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr7_+_12144162 | 0.04 |
ENSRNOT00000077122
|
Tcf3
|
transcription factor 3 |
| chr10_-_47857326 | 0.04 |
ENSRNOT00000085703
ENSRNOT00000091963 |
Epn2
|
epsin 2 |
| chr5_-_122746839 | 0.04 |
ENSRNOT00000032126
|
Slc35d1
|
solute carrier family 35 member D1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.3 | GO:1902380 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.1 | 0.6 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.4 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.2 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.1 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) |
| 0.0 | 0.1 | GO:0097213 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 0.0 | 0.2 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) positive regulation of dermatome development(GO:0061184) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0021979 | forebrain ventricular zone progenitor cell division(GO:0021869) hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0070212 | regulation of chromatin assembly(GO:0010847) protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.0 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.0 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.0 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.3 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.1 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |