Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for E2f8
Z-value: 0.26
Transcription factors associated with E2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f8
|
ENSRNOG00000022537 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f8 | rn6_v1_chr1_-_104202591_104202591 | -0.67 | 2.2e-01 | Click! |
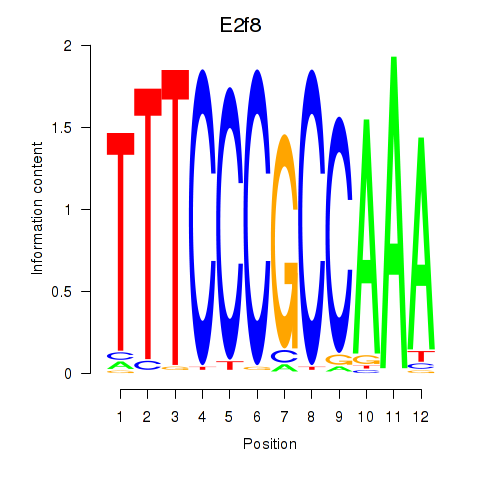
Activity profile of E2f8 motif
Sorted Z-values of E2f8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16395029 | 0.10 |
ENSRNOT00000001700
|
Nudt1
|
nudix hydrolase 1 |
| chr11_-_83867203 | 0.07 |
ENSRNOT00000002394
|
Chrd
|
chordin |
| chr1_+_221558093 | 0.06 |
ENSRNOT00000077274
|
Majin
|
membrane anchored junction protein |
| chr19_+_14523554 | 0.06 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr4_-_179904844 | 0.05 |
ENSRNOT00000085386
|
Lmntd1
|
lamin tail domain containing 1 |
| chr14_+_110676090 | 0.04 |
ENSRNOT00000029513
|
Fancl
|
Fanconi anemia, complementation group L |
| chr4_+_157554794 | 0.04 |
ENSRNOT00000024116
|
Ing4
|
inhibitor of growth family, member 4 |
| chr10_-_89699836 | 0.04 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr8_-_39201588 | 0.04 |
ENSRNOT00000011226
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr3_-_80052953 | 0.04 |
ENSRNOT00000018998
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr2_+_197682000 | 0.03 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr1_+_185863043 | 0.03 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr2_-_28423762 | 0.03 |
ENSRNOT00000022864
|
Btf3
|
basic transcription factor 3 |
| chr9_-_61975640 | 0.03 |
ENSRNOT00000085744
|
Boll
|
boule homolog, RNA binding protein |
| chr1_-_32362359 | 0.03 |
ENSRNOT00000079987
|
Slc6a3
|
solute carrier family 6 member 3 |
| chr9_-_64573660 | 0.03 |
ENSRNOT00000021299
|
LOC108348134
|
protein boule-like |
| chr11_-_57993548 | 0.03 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr1_+_220009397 | 0.03 |
ENSRNOT00000084410
|
Rbm4b
|
RNA binding motif protein 4B |
| chr5_+_154522119 | 0.03 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr3_-_150073721 | 0.03 |
ENSRNOT00000022428
|
E2f1
|
E2F transcription factor 1 |
| chr8_+_108958046 | 0.02 |
ENSRNOT00000079618
ENSRNOT00000066917 |
Stag1
|
stromal antigen 1 |
| chr4_+_62703779 | 0.02 |
ENSRNOT00000014832
|
Nup205
|
nucleoporin 205 |
| chr10_+_90622992 | 0.02 |
ENSRNOT00000032856
|
Meioc
|
meiosis specific with coiled-coil domain |
| chr3_+_2158995 | 0.02 |
ENSRNOT00000010270
|
Zmynd19
|
zinc finger, MYND-type containing 19 |
| chr13_+_48679774 | 0.02 |
ENSRNOT00000075674
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr14_+_82356916 | 0.02 |
ENSRNOT00000040229
|
Slbp
|
stem-loop binding protein |
| chr7_+_54859104 | 0.02 |
ENSRNOT00000087341
|
Caps2
|
calcyphosine 2 |
| chr4_+_179905116 | 0.02 |
ENSRNOT00000052352
|
Tuba3a
|
tubulin, alpha 3A |
| chr1_+_257901985 | 0.02 |
ENSRNOT00000073015
|
Hells
|
helicase, lymphoid specific |
| chr6_+_43884678 | 0.02 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr11_-_86890390 | 0.02 |
ENSRNOT00000046183
|
Trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr1_+_257766691 | 0.02 |
ENSRNOT00000088148
|
Hells
|
helicase, lymphoid specific |
| chr10_+_93354003 | 0.02 |
ENSRNOT00000008140
|
Mettl2b
|
methyltransferase like 2B |
| chr1_+_101687855 | 0.02 |
ENSRNOT00000028546
ENSRNOT00000091597 |
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr3_-_160891190 | 0.02 |
ENSRNOT00000019386
|
Sdc4
|
syndecan 4 |
| chr10_-_56839437 | 0.02 |
ENSRNOT00000090338
|
Bcl6b
|
B-cell CLL/lymphoma 6B |
| chr1_+_257614731 | 0.02 |
ENSRNOT00000075680
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr3_+_159421671 | 0.02 |
ENSRNOT00000010343
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr14_+_106393959 | 0.01 |
ENSRNOT00000092168
|
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr3_-_151486693 | 0.01 |
ENSRNOT00000073736
ENSRNOT00000071099 |
Gdf5
|
growth differentiation factor 5 |
| chr11_+_86890585 | 0.01 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr6_+_95816749 | 0.01 |
ENSRNOT00000008880
|
Six6
|
SIX homeobox 6 |
| chr2_+_186630278 | 0.01 |
ENSRNOT00000021604
|
LOC365839
|
similar to elongation protein 4 homolog |
| chr14_-_106393670 | 0.01 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chr20_+_25990656 | 0.01 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr2_-_259898525 | 0.01 |
ENSRNOT00000082253
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr7_+_54859326 | 0.01 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr11_-_89510871 | 0.01 |
ENSRNOT00000035247
|
Prkdc
|
protein kinase, DNA activated, catalytic polypeptide |
| chr9_-_10757954 | 0.01 |
ENSRNOT00000075265
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr2_+_24546536 | 0.01 |
ENSRNOT00000014036
|
Otp
|
orthopedia homeobox |
| chr18_+_30036887 | 0.01 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr10_+_75335146 | 0.01 |
ENSRNOT00000073005
|
Srsf1
|
serine and arginine rich splicing factor 1 |
| chr9_-_10757720 | 0.01 |
ENSRNOT00000083848
|
Uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr19_-_44231157 | 0.01 |
ENSRNOT00000046126
|
Kars
|
lysyl-tRNA synthetase |
| chr1_+_257615088 | 0.01 |
ENSRNOT00000083183
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chrX_-_112159458 | 0.01 |
ENSRNOT00000087403
|
Tex13b
|
testis expressed 13B |
| chr6_+_128973956 | 0.01 |
ENSRNOT00000075399
|
Fam181a
|
family with sequence similarity 181, member A |
| chr8_-_55408464 | 0.00 |
ENSRNOT00000066893
|
Sik2
|
salt-inducible kinase 2 |
| chr5_+_147185474 | 0.00 |
ENSRNOT00000000134
|
Ak2
|
adenylate kinase 2 |
| chr14_+_44580216 | 0.00 |
ENSRNOT00000088674
ENSRNOT00000003907 |
Rfc1
|
replication factor C subunit 1 |
| chr1_+_174767960 | 0.00 |
ENSRNOT00000013362
|
Wee1
|
WEE1 G2 checkpoint kinase |
| chr14_+_48726045 | 0.00 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_+_147375350 | 0.00 |
ENSRNOT00000010674
|
Yars
|
tyrosyl-tRNA synthetase |
| chr19_+_37652969 | 0.00 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr18_+_56364620 | 0.00 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr2_-_231881939 | 0.00 |
ENSRNOT00000074644
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr6_+_76675418 | 0.00 |
ENSRNOT00000076169
ENSRNOT00000010948 ENSRNOT00000082286 |
Brms1l
Brms1l
|
breast cancer metastasis-suppressor 1-like breast cancer metastasis-suppressor 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |