Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for E2f7
Z-value: 0.17
Transcription factors associated with E2f7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f7
|
ENSRNOG00000026252 | E2F transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f7 | rn6_v1_chr7_+_53275676_53275676 | -0.05 | 9.4e-01 | Click! |
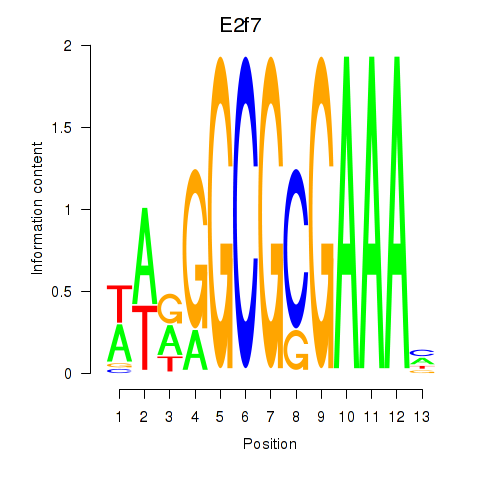
Activity profile of E2f7 motif
Sorted Z-values of E2f7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_44793927 | 0.06 |
ENSRNOT00000086309
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr3_+_103747654 | 0.05 |
ENSRNOT00000006887
|
Nop10
|
NOP10 ribonucleoprotein |
| chr8_+_59344083 | 0.04 |
ENSRNOT00000031175
|
Crabp1
|
cellular retinoic acid binding protein 1 |
| chr17_+_44794130 | 0.04 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr17_-_44520240 | 0.03 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr10_+_62650719 | 0.03 |
ENSRNOT00000010691
|
Tp53i13
|
tumor protein p53 inducible protein 13 |
| chr12_+_22665112 | 0.03 |
ENSRNOT00000001918
|
Ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr17_+_44763598 | 0.03 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr13_-_73390393 | 0.03 |
ENSRNOT00000067253
ENSRNOT00000093438 |
Lhx4
|
LIM homeobox 4 |
| chr3_+_59153280 | 0.03 |
ENSRNOT00000002066
|
Cdca7
|
cell division cycle associated 7 |
| chr17_-_44815995 | 0.03 |
ENSRNOT00000091201
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr10_-_57653359 | 0.03 |
ENSRNOT00000089638
|
Derl2
|
derlin 2 |
| chr11_+_86890585 | 0.02 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr1_+_80056755 | 0.02 |
ENSRNOT00000021221
|
Snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr17_+_9830332 | 0.02 |
ENSRNOT00000021946
|
Rab24
|
RAB24, member RAS oncogene family |
| chr1_-_84986581 | 0.02 |
ENSRNOT00000025819
|
Psmc4
|
proteasome 26S subunit, ATPase 4 |
| chr1_+_80000165 | 0.02 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr5_-_21345805 | 0.02 |
ENSRNOT00000081296
ENSRNOT00000007802 |
Car8
|
carbonic anhydrase 8 |
| chr13_-_109849632 | 0.02 |
ENSRNOT00000005085
|
Atf3
|
activating transcription factor 3 |
| chr1_-_48033343 | 0.02 |
ENSRNOT00000019531
ENSRNOT00000076422 |
Tcp1
|
t-complex 1 |
| chr10_-_6870011 | 0.02 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr10_+_3218466 | 0.02 |
ENSRNOT00000093629
ENSRNOT00000093338 ENSRNOT00000077695 |
Ntan1
|
N-terminal asparagine amidase |
| chr10_+_13786459 | 0.02 |
ENSRNOT00000010915
|
Rnps1
|
RNA binding protein with serine rich domain 1 |
| chr10_+_101288489 | 0.02 |
ENSRNOT00000003511
|
Sox9
|
SRY box 9 |
| chr1_+_48033768 | 0.02 |
ENSRNOT00000020064
|
Mrpl18
|
mitochondrial ribosomal protein L18 |
| chr14_-_86706626 | 0.01 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr2_-_188413219 | 0.01 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chr3_-_176431423 | 0.01 |
ENSRNOT00000037271
|
Ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr17_-_43798383 | 0.01 |
ENSRNOT00000075069
|
LOC684828
|
similar to Histone H1.2 (H1 VAR.1) (H1c) |
| chr6_+_37144787 | 0.01 |
ENSRNOT00000075503
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr2_+_187893368 | 0.01 |
ENSRNOT00000092031
|
Mex3a
|
mex-3 RNA binding family member A |
| chr2_-_250232295 | 0.01 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr9_-_100888107 | 0.01 |
ENSRNOT00000024794
|
Thap4
|
THAP domain containing 4 |
| chr20_+_4530342 | 0.01 |
ENSRNOT00000076352
ENSRNOT00000000478 ENSRNOT00000075925 |
Nelfe
|
negative elongation factor complex member E |
| chr8_+_119135013 | 0.01 |
ENSRNOT00000056114
|
Prss50
|
protease, serine, 50 |
| chr17_+_44738643 | 0.01 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr15_-_42518855 | 0.01 |
ENSRNOT00000076451
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_+_44528125 | 0.01 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr1_-_228263198 | 0.01 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr11_-_89260297 | 0.01 |
ENSRNOT00000057502
|
Spidr
|
scaffolding protein involved in DNA repair |
| chr16_-_6404578 | 0.01 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr3_+_114129589 | 0.01 |
ENSRNOT00000056119
|
Terb2
|
telomere repeat binding bouquet formation protein 2 |
| chr11_+_82373870 | 0.01 |
ENSRNOT00000002429
|
Tra2b
|
transformer 2 beta homolog (Drosophila) |
| chr17_-_44841382 | 0.01 |
ENSRNOT00000080119
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr3_-_91217491 | 0.01 |
ENSRNOT00000006115
|
Rag1
|
recombination activating 1 |
| chr1_-_142759882 | 0.01 |
ENSRNOT00000015218
|
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr1_+_145770135 | 0.01 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr2_-_250778269 | 0.01 |
ENSRNOT00000085670
ENSRNOT00000083535 ENSRNOT00000017824 |
Clca5
|
chloride channel accessory 5 |
| chr10_-_108150511 | 0.01 |
ENSRNOT00000073337
|
Cbx8
|
chromobox 8 |
| chr6_-_38228379 | 0.01 |
ENSRNOT00000084924
|
Mycn
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr4_-_157679962 | 0.01 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr20_-_5037022 | 0.01 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr1_+_193335645 | 0.01 |
ENSRNOT00000019640
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr1_-_228226354 | 0.01 |
ENSRNOT00000029224
|
Olr318
|
olfactory receptor 318 |
| chr3_-_83048289 | 0.01 |
ENSRNOT00000047571
ENSRNOT00000012806 |
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr1_-_82546937 | 0.01 |
ENSRNOT00000028100
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr5_-_150609352 | 0.01 |
ENSRNOT00000074374
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr7_-_95310005 | 0.01 |
ENSRNOT00000005815
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr1_-_80056574 | 0.01 |
ENSRNOT00000021200
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr6_+_43884678 | 0.01 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr10_-_103736973 | 0.01 |
ENSRNOT00000004372
|
Nat9
|
N-acetyltransferase 9 |
| chr7_+_123531682 | 0.01 |
ENSRNOT00000010606
|
Wbp2nl
|
WBP2 N-terminal like |
| chr4_+_57823411 | 0.01 |
ENSRNOT00000030462
ENSRNOT00000088235 |
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr8_+_48422036 | 0.01 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr3_-_124145566 | 0.01 |
ENSRNOT00000028877
|
Adra1d
|
adrenoceptor alpha 1D |
| chr13_-_80703615 | 0.01 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr8_+_39960542 | 0.01 |
ENSRNOT00000013370
|
Msantd2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr19_+_14523554 | 0.00 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr6_+_10912383 | 0.00 |
ENSRNOT00000061747
ENSRNOT00000086247 |
Ttc7a
|
tetratricopeptide repeat domain 7A |
| chr6_+_36941233 | 0.00 |
ENSRNOT00000007073
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr10_+_105235289 | 0.00 |
ENSRNOT00000075548
|
Galr2
|
galanin receptor 2 |
| chr10_-_78993045 | 0.00 |
ENSRNOT00000043005
|
LOC100363469
|
ribosomal protein S24-like |
| chr4_-_144318580 | 0.00 |
ENSRNOT00000007591
|
Ssuh2
|
ssu-2 homolog |
| chr5_+_154205402 | 0.00 |
ENSRNOT00000064925
|
Srsf10
|
serine and arginine rich splicing factor 10 |
| chr8_+_108958046 | 0.00 |
ENSRNOT00000079618
ENSRNOT00000066917 |
Stag1
|
stromal antigen 1 |
| chr20_-_3298835 | 0.00 |
ENSRNOT00000083617
|
Gnl1
|
G protein nucleolar 1 |
| chr2_-_188471988 | 0.00 |
ENSRNOT00000027785
|
Hcn3
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 3 |
| chr13_+_90909486 | 0.00 |
ENSRNOT00000011450
|
Cfap45
|
cilia and flagella associated protein 45 |
| chr10_-_46206135 | 0.00 |
ENSRNOT00000091471
|
Cops3
|
COP9 signalosome subunit 3 |
| chr7_+_12192088 | 0.00 |
ENSRNOT00000040170
|
Mex3d
|
mex-3 RNA binding family member D |
| chr20_+_19318250 | 0.00 |
ENSRNOT00000000299
|
Phyhipl
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr1_-_226935689 | 0.00 |
ENSRNOT00000038807
|
Tmem109
|
transmembrane protein 109 |
| chr1_+_174767960 | 0.00 |
ENSRNOT00000013362
|
Wee1
|
WEE1 G2 checkpoint kinase |
| chr6_+_76675418 | 0.00 |
ENSRNOT00000076169
ENSRNOT00000010948 ENSRNOT00000082286 |
Brms1l
Brms1l
|
breast cancer metastasis-suppressor 1-like breast cancer metastasis-suppressor 1-like |
| chr9_+_81783349 | 0.00 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr6_+_36941596 | 0.00 |
ENSRNOT00000083383
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr1_+_228337767 | 0.00 |
ENSRNOT00000066247
|
Patl1
|
PAT1 homolog 1, processing body mRNA decay factor |
| chrX_+_54734385 | 0.00 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr17_-_44527801 | 0.00 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |