Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
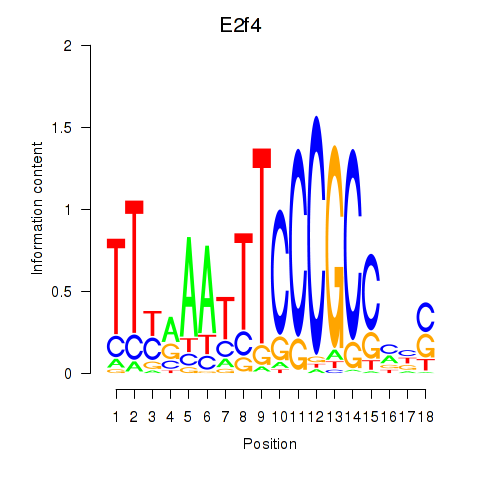
Results for E2f4
Z-value: 0.29
Transcription factors associated with E2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f4
|
ENSRNOG00000015708 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | rn6_v1_chr19_+_37252843_37252843 | -0.98 | 2.4e-03 | Click! |
Activity profile of E2f4 motif
Sorted Z-values of E2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_28715224 | 0.22 |
ENSRNOT00000065899
|
Parpbp
|
PARP1 binding protein |
| chr15_-_61564695 | 0.14 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr9_-_82146874 | 0.11 |
ENSRNOT00000024190
|
Fev
|
FEV, ETS transcription factor |
| chr13_+_75059927 | 0.09 |
ENSRNOT00000080801
|
LOC680254
|
hypothetical protein LOC680254 |
| chr2_-_187915596 | 0.09 |
ENSRNOT00000093008
|
Lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr2_+_187447501 | 0.08 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr20_+_4967194 | 0.08 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr10_+_55626741 | 0.08 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr5_-_152198813 | 0.08 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_87435185 | 0.08 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr5_-_57267002 | 0.08 |
ENSRNOT00000011455
|
Bag1
|
Bcl2 associated athanogene 1 |
| chr19_-_55967836 | 0.07 |
ENSRNOT00000051452
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr10_-_15098791 | 0.07 |
ENSRNOT00000026139
|
Chtf18
|
chromosome transmission fidelity factor 18 |
| chr20_-_7930929 | 0.07 |
ENSRNOT00000000607
|
Tead3
|
TEA domain transcription factor 3 |
| chr5_+_103024376 | 0.07 |
ENSRNOT00000058536
|
AABR07049064.1
|
|
| chr1_+_31967978 | 0.07 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr16_+_41079444 | 0.07 |
ENSRNOT00000015623
|
Neil3
|
nei-like DNA glycosylase 3 |
| chr5_+_162351001 | 0.07 |
ENSRNOT00000065471
|
LOC691162
|
hypothetical protein LOC691162 |
| chr12_+_943006 | 0.06 |
ENSRNOT00000001449
|
Kl
|
Klotho |
| chr8_-_12993651 | 0.06 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr9_+_16139101 | 0.06 |
ENSRNOT00000070803
|
LOC100910668
|
uncharacterized LOC100910668 |
| chr6_+_10594122 | 0.06 |
ENSRNOT00000020534
|
Cript
|
CXXC repeat containing interactor of PDZ3 domain |
| chr13_+_110743098 | 0.06 |
ENSRNOT00000075086
|
Rd3
|
retinal degeneration 3 |
| chr9_+_9842585 | 0.05 |
ENSRNOT00000070806
|
Cd70
|
Cd70 molecule |
| chr10_+_55627025 | 0.05 |
ENSRNOT00000091016
|
Aurkb
|
aurora kinase B |
| chr14_-_110883784 | 0.05 |
ENSRNOT00000086299
ENSRNOT00000068190 |
Vrk2
|
vaccinia related kinase 2 |
| chr1_-_89488223 | 0.05 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr11_-_36479868 | 0.05 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr7_+_120901934 | 0.05 |
ENSRNOT00000019323
|
Tomm22
|
translocase of outer mitochondrial membrane 22 |
| chr6_+_34155017 | 0.05 |
ENSRNOT00000073663
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr20_-_3439983 | 0.04 |
ENSRNOT00000080822
ENSRNOT00000001099 |
Ier3
|
immediate early response 3 |
| chr8_-_52814188 | 0.04 |
ENSRNOT00000087265
ENSRNOT00000047633 |
Rexo2
|
RNA exonuclease 2 |
| chr6_-_7421456 | 0.04 |
ENSRNOT00000006725
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr3_+_159421671 | 0.04 |
ENSRNOT00000010343
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_+_7258176 | 0.04 |
ENSRNOT00000061992
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr10_+_39726507 | 0.04 |
ENSRNOT00000036822
|
Meikin
|
meiotic kinetochore factor |
| chr8_+_48665652 | 0.04 |
ENSRNOT00000059715
|
H2afx
|
H2A histone family, member X |
| chr20_-_6500523 | 0.04 |
ENSRNOT00000000629
|
Cpne5
|
copine 5 |
| chr2_+_264704769 | 0.04 |
ENSRNOT00000012667
|
Depdc1
|
DEP domain containing 1 |
| chr17_-_78735324 | 0.04 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr14_+_86813082 | 0.04 |
ENSRNOT00000090360
|
Ccm2
|
CCM2 scaffolding protein |
| chr5_-_105212173 | 0.04 |
ENSRNOT00000010383
|
Rps6
|
ribosomal protein S6 |
| chr5_+_128450680 | 0.04 |
ENSRNOT00000010700
|
Txndc12
|
thioredoxin domain containing 12 |
| chr20_-_5037022 | 0.04 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr15_-_60512704 | 0.04 |
ENSRNOT00000049056
|
Tnfsf11
|
tumor necrosis factor superfamily member 11 |
| chr8_+_79054237 | 0.04 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr13_-_55886679 | 0.03 |
ENSRNOT00000066601
|
RGD1566099
|
similar to novel protein |
| chr1_+_48433079 | 0.03 |
ENSRNOT00000037369
|
Slc22a3
|
solute carrier family 22 member 3 |
| chr10_-_63499390 | 0.03 |
ENSRNOT00000032934
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr4_+_7257752 | 0.03 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr6_-_55647665 | 0.03 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chrX_-_123731294 | 0.03 |
ENSRNOT00000092574
ENSRNOT00000032618 |
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr15_-_70399924 | 0.03 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr10_-_86393141 | 0.03 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr9_-_16807966 | 0.03 |
ENSRNOT00000073160
|
Rps19
|
ribosomal protein S19 |
| chr1_+_221448661 | 0.03 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chr1_+_81750928 | 0.03 |
ENSRNOT00000027246
|
NEWGENE_68440
|
ribosomal protein S19 |
| chr4_-_7281223 | 0.03 |
ENSRNOT00000019666
|
Slc4a2
|
solute carrier family 4 member 2 |
| chr5_+_57267312 | 0.03 |
ENSRNOT00000011907
|
Chmp5
|
charged multivesicular body protein 5 |
| chrX_-_158655198 | 0.03 |
ENSRNOT00000045153
|
LOC103689983
|
hypoxanthine-guanine phosphoribosyltransferase |
| chr10_+_4313100 | 0.03 |
ENSRNOT00000074487
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr1_-_1116926 | 0.03 |
ENSRNOT00000062027
|
Raet1l
|
retinoic acid early transcript 1L |
| chr20_+_4966817 | 0.03 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_+_81566074 | 0.03 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr8_+_12994155 | 0.02 |
ENSRNOT00000011855
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr4_-_160803558 | 0.02 |
ENSRNOT00000049542
|
Senp18
|
Sumo1/sentrin/SMT3 specific peptidase 18 |
| chr6_+_129835919 | 0.02 |
ENSRNOT00000036035
|
Vrk1
|
vaccinia related kinase 1 |
| chr3_+_91463660 | 0.02 |
ENSRNOT00000081984
ENSRNOT00000006628 |
Commd9
|
COMM domain containing 9 |
| chr9_+_64066579 | 0.02 |
ENSRNOT00000013511
|
RGD1306941
|
similar to CG31122-PA |
| chr6_+_55648021 | 0.02 |
ENSRNOT00000064822
ENSRNOT00000091488 |
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr7_-_140951071 | 0.02 |
ENSRNOT00000082521
|
Fam186b
|
family with sequence similarity 186, member B |
| chr10_-_45851984 | 0.02 |
ENSRNOT00000058308
|
Zfp496
|
zinc finger protein 496 |
| chr1_-_221448570 | 0.02 |
ENSRNOT00000075165
|
Zfpl1
|
zinc finger protein-like 1 |
| chr20_+_27169267 | 0.02 |
ENSRNOT00000077525
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr2_+_244521699 | 0.02 |
ENSRNOT00000029382
|
Stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chrX_-_158228749 | 0.02 |
ENSRNOT00000065935
|
Hprt1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr14_-_33580566 | 0.02 |
ENSRNOT00000063942
|
Paics
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr4_-_145487426 | 0.02 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr15_-_24199341 | 0.02 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr12_+_48101673 | 0.02 |
ENSRNOT00000065060
|
Foxn4
|
forkhead box N4 |
| chr15_-_42638392 | 0.01 |
ENSRNOT00000022020
|
Scara3
|
scavenger receptor class A, member 3 |
| chr6_-_43862131 | 0.01 |
ENSRNOT00000089859
|
Cys1
|
cystin 1 |
| chr13_-_108178609 | 0.01 |
ENSRNOT00000004525
|
Cenpf
|
centromere protein F |
| chr8_+_44847157 | 0.01 |
ENSRNOT00000080288
|
Clmp
|
CXADR-like membrane protein |
| chr8_-_65587658 | 0.01 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr9_-_9934501 | 0.01 |
ENSRNOT00000074904
|
AABR07066510.1
|
|
| chr3_+_111597102 | 0.01 |
ENSRNOT00000081462
|
Tyro3
|
TYRO3 protein tyrosine kinase |
| chr9_-_81566096 | 0.01 |
ENSRNOT00000019497
|
Aamp
|
angio-associated, migratory cell protein |
| chr16_+_19800463 | 0.01 |
ENSRNOT00000023065
|
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr3_+_176217128 | 0.01 |
ENSRNOT00000013609
|
Gid8
|
GID complex subunit 8 |
| chr13_+_70852023 | 0.01 |
ENSRNOT00000003661
|
Shcbp1l
|
SHC binding and spindle associated 1 like |
| chr4_-_10329241 | 0.01 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr10_+_4312863 | 0.01 |
ENSRNOT00000091610
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr12_-_19314016 | 0.01 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr1_-_198652674 | 0.01 |
ENSRNOT00000023706
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr8_-_132032971 | 0.01 |
ENSRNOT00000005502
|
RGD1311745
|
similar to RIKEN cDNA 1110059G10 |
| chr1_-_82546937 | 0.01 |
ENSRNOT00000028100
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr11_-_74866451 | 0.01 |
ENSRNOT00000002330
|
Il13ra1
|
interleukin 13 receptor subunit alpha 1 |
| chr3_+_55960327 | 0.01 |
ENSRNOT00000086584
|
Ppig
|
peptidylprolyl isomerase G |
| chr10_-_89454681 | 0.01 |
ENSRNOT00000028109
|
Brca1
|
BRCA1, DNA repair associated |
| chr9_-_92963697 | 0.01 |
ENSRNOT00000023546
|
Gpr55
|
G protein-coupled receptor 55 |
| chr1_-_184106200 | 0.01 |
ENSRNOT00000067609
|
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr8_+_70860671 | 0.01 |
ENSRNOT00000019258
|
Pdcd7
|
programmed cell death 7 |
| chrX_+_15171499 | 0.01 |
ENSRNOT00000008399
|
Suv39h1l1
|
suppressor of variegation 3-9 homolog 1 (Drosophila)-like 1 |
| chr5_-_151029233 | 0.01 |
ENSRNOT00000089155
|
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
| chr5_+_152721940 | 0.01 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chrX_-_62903530 | 0.01 |
ENSRNOT00000076652
ENSRNOT00000017370 |
Pdk3
|
pyruvate dehydrogenase kinase 3 |
| chr17_+_78735598 | 0.01 |
ENSRNOT00000020854
|
Hspa14
|
heat shock protein family A, member 14 |
| chr1_-_198232344 | 0.01 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr3_+_55960067 | 0.01 |
ENSRNOT00000010216
|
Ppig
|
peptidylprolyl isomerase G |
| chr1_+_85470831 | 0.01 |
ENSRNOT00000057096
|
Timm50
|
translocase of inner mitochondrial membrane 50 |
| chr19_+_10024947 | 0.01 |
ENSRNOT00000061392
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr4_-_56897310 | 0.01 |
ENSRNOT00000043902
ENSRNOT00000090038 |
Tnpo3
|
transportin 3 |
| chr13_-_87847263 | 0.01 |
ENSRNOT00000003650
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr10_+_10811937 | 0.01 |
ENSRNOT00000077347
|
Anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr8_-_49269411 | 0.01 |
ENSRNOT00000021321
|
Ube4a
|
ubiquitination factor E4A |
| chr9_+_93326283 | 0.01 |
ENSRNOT00000024578
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr1_-_157700064 | 0.01 |
ENSRNOT00000031974
|
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr3_+_1478525 | 0.01 |
ENSRNOT00000008161
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr1_-_94494980 | 0.00 |
ENSRNOT00000020014
|
Ccne1
|
cyclin E1 |
| chrX_+_15295473 | 0.00 |
ENSRNOT00000009295
|
LOC108348065
|
histone deacetylase 6 |
| chr8_-_65587427 | 0.00 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr8_-_69807826 | 0.00 |
ENSRNOT00000014071
|
Dis3l
|
DIS3-like exosome 3'-5' exoribonuclease |
| chr1_-_31967915 | 0.00 |
ENSRNOT00000021000
|
Brd9
|
bromodomain containing 9 |
| chr5_+_154205402 | 0.00 |
ENSRNOT00000064925
|
Srsf10
|
serine and arginine rich splicing factor 10 |
| chr2_-_208420163 | 0.00 |
ENSRNOT00000021920
|
LOC100911417
|
ATP synthase subunit b, mitochondrial-like |
| chr5_+_76860515 | 0.00 |
ENSRNOT00000022866
|
RGD1310951
|
similar to RIKEN cDNA E130308A19 |
| chr14_+_82356916 | 0.00 |
ENSRNOT00000040229
|
Slbp
|
stem-loop binding protein |
| chr10_+_4411766 | 0.00 |
ENSRNOT00000003213
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr8_-_111850393 | 0.00 |
ENSRNOT00000044956
|
Cdv3
|
CDV3 homolog |
| chr6_+_10306405 | 0.00 |
ENSRNOT00000090420
ENSRNOT00000080507 |
Epas1
|
endothelial PAS domain protein 1 |
| chr3_+_149131785 | 0.00 |
ENSRNOT00000090321
|
Dnmt3b
|
DNA methyltransferase 3 beta |
| chr4_-_52350624 | 0.00 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
| chr7_+_28715300 | 0.00 |
ENSRNOT00000006760
ENSRNOT00000089161 |
Nup37
|
nucleoporin 37 |
| chr2_+_187915751 | 0.00 |
ENSRNOT00000026994
|
Ubqln4
|
ubiquilin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.0 | GO:0006178 | guanine salvage(GO:0006178) GMP catabolic process(GO:0046038) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |