Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
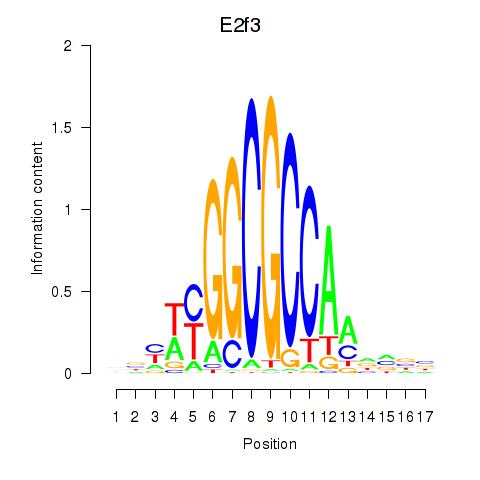
Results for E2f3
Z-value: 0.36
Transcription factors associated with E2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f3
|
ENSRNOG00000029273 | E2F transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f3 | rn6_v1_chr17_+_36334589_36334589 | 0.37 | 5.4e-01 | Click! |
Activity profile of E2f3 motif
Sorted Z-values of E2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_943006 | 0.25 |
ENSRNOT00000001449
|
Kl
|
Klotho |
| chr15_-_30147793 | 0.13 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr1_-_219422268 | 0.10 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chr20_-_7654428 | 0.10 |
ENSRNOT00000045044
|
Tcp11
|
t-complex 11 |
| chr2_+_30664639 | 0.09 |
ENSRNOT00000076372
ENSRNOT00000076294 ENSRNOT00000076434 ENSRNOT00000076484 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr20_-_14282873 | 0.09 |
ENSRNOT00000001759
|
Adora2a
|
adenosine A2a receptor |
| chr1_-_42467586 | 0.09 |
ENSRNOT00000029416
|
Fbxo5
|
F-box protein 5 |
| chr10_-_53037816 | 0.08 |
ENSRNOT00000057509
|
Shisa6
|
shisa family member 6 |
| chr13_-_45068077 | 0.08 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr4_-_152380023 | 0.08 |
ENSRNOT00000012397
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr6_+_33885495 | 0.08 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr5_+_47853818 | 0.07 |
ENSRNOT00000009228
ENSRNOT00000079656 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr2_+_198231291 | 0.07 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr2_+_4942775 | 0.06 |
ENSRNOT00000093548
ENSRNOT00000093741 |
Fam172a
|
family with sequence similarity 172, member A |
| chr1_-_270472866 | 0.06 |
ENSRNOT00000015979
|
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr8_-_122402127 | 0.06 |
ENSRNOT00000074238
ENSRNOT00000076548 |
Crtapl1
|
cartilage associated protein-like 1 |
| chr11_+_73198522 | 0.06 |
ENSRNOT00000002356
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr11_-_51202703 | 0.06 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr8_+_69971778 | 0.06 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr18_-_47513030 | 0.05 |
ENSRNOT00000083881
ENSRNOT00000074226 |
Lox
|
lysyl oxidase |
| chr8_-_118378460 | 0.05 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr3_+_138715570 | 0.05 |
ENSRNOT00000064723
|
Sec23b
|
Sec23 homolog B, coat complex II component |
| chr9_+_121802673 | 0.05 |
ENSRNOT00000086534
|
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr1_+_165724451 | 0.05 |
ENSRNOT00000025827
|
Fam168a
|
family with sequence similarity 168, member A |
| chr6_-_1622196 | 0.05 |
ENSRNOT00000007492
|
Prkd3
|
protein kinase D3 |
| chr3_-_2938883 | 0.05 |
ENSRNOT00000084196
|
LOC108348105
|
allergen Fel d 4-like |
| chr7_-_24667301 | 0.05 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chr8_+_117953444 | 0.05 |
ENSRNOT00000028141
|
Cdc25a
|
cell division cycle 25A |
| chr13_+_110257571 | 0.05 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr4_-_78342863 | 0.05 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr7_-_124198200 | 0.05 |
ENSRNOT00000078947
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr5_-_169331163 | 0.05 |
ENSRNOT00000042301
|
Espn
|
espin |
| chr12_-_47793534 | 0.05 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr3_-_55951584 | 0.05 |
ENSRNOT00000036585
|
Fastkd1
|
FAST kinase domains 1 |
| chr19_+_52521809 | 0.05 |
ENSRNOT00000081019
|
Klhl36
|
kelch-like family member 36 |
| chr18_+_30550877 | 0.05 |
ENSRNOT00000027164
|
LOC108348201
|
protocadherin beta-7-like |
| chr9_-_46206605 | 0.04 |
ENSRNOT00000018640
|
Tbc1d8
|
TBC1 domain family, member 8 |
| chr17_-_20364714 | 0.04 |
ENSRNOT00000070962
|
Jarid2
|
jumonji and AT-rich interaction domain containing 2 |
| chr3_-_3660006 | 0.04 |
ENSRNOT00000045587
|
Lhx3
|
LIM homeobox 3 |
| chr3_+_12262822 | 0.04 |
ENSRNOT00000022585
|
Angptl2
|
angiopoietin-like 2 |
| chr3_+_146582752 | 0.04 |
ENSRNOT00000010157
|
Pygb
|
glycogen phosphorylase B |
| chr8_+_22446661 | 0.04 |
ENSRNOT00000010030
|
Qtrt1
|
queuine tRNA-ribosyltransferase catalytic subunit 1 |
| chr4_+_157438605 | 0.04 |
ENSRNOT00000079988
|
AC115420.4
|
|
| chr7_+_130474279 | 0.04 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr8_-_87245774 | 0.04 |
ENSRNOT00000085725
|
Tmem30a
|
transmembrane protein 30A |
| chr18_-_38088457 | 0.04 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr8_+_7128656 | 0.04 |
ENSRNOT00000038313
|
Pgr
|
progesterone receptor |
| chr10_-_70118194 | 0.04 |
ENSRNOT00000009944
|
Cct6b
|
chaperonin containing TCP1 subunit 6B |
| chr8_+_48422036 | 0.04 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr2_-_157759819 | 0.04 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr11_-_68842320 | 0.04 |
ENSRNOT00000049180
|
Adcy5
|
adenylate cyclase 5 |
| chrX_+_74200972 | 0.04 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr7_+_73588163 | 0.04 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr2_-_198412350 | 0.04 |
ENSRNOT00000040210
|
LOC100912489
|
histone H4-like |
| chr13_+_74154835 | 0.04 |
ENSRNOT00000059524
|
Abl2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr1_+_126749508 | 0.04 |
ENSRNOT00000015845
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr11_-_27080701 | 0.04 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr1_-_1885981 | 0.04 |
ENSRNOT00000020810
|
Ginm1
|
glycoprotein integral membrane 1 |
| chr5_+_138470069 | 0.04 |
ENSRNOT00000076343
ENSRNOT00000064073 |
Zmynd12
|
zinc finger, MYND-type containing 12 |
| chr6_-_15191660 | 0.04 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr6_-_91518996 | 0.04 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr15_+_34270648 | 0.03 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr6_-_124735741 | 0.03 |
ENSRNOT00000064716
ENSRNOT00000091693 |
Rps6ka5
|
ribosomal protein S6 kinase A5 |
| chr16_-_5795825 | 0.03 |
ENSRNOT00000048043
|
Cacna2d3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3 |
| chr10_+_82292110 | 0.03 |
ENSRNOT00000004435
|
Chad
|
chondroadherin |
| chr1_+_222746023 | 0.03 |
ENSRNOT00000028787
|
Atl3
|
atlastin GTPase 3 |
| chr9_-_46309451 | 0.03 |
ENSRNOT00000018684
|
Rnf149
|
ring finger protein 149 |
| chr9_+_10471742 | 0.03 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr7_+_117963740 | 0.03 |
ENSRNOT00000075405
|
LOC108348189
|
COMM domain-containing protein 5 |
| chr20_+_48504264 | 0.03 |
ENSRNOT00000087740
|
Cdc40
|
cell division cycle 40 |
| chr2_+_128675814 | 0.03 |
ENSRNOT00000058366
|
RGD1359508
|
similar to protein C33A12.3 |
| chr6_+_119519714 | 0.03 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr13_+_44475970 | 0.03 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr6_-_95890325 | 0.03 |
ENSRNOT00000050217
|
LOC690335
|
similar to 60S ribosomal protein L23a |
| chr8_-_48619592 | 0.03 |
ENSRNOT00000012534
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chr12_+_660011 | 0.03 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr5_+_128923934 | 0.03 |
ENSRNOT00000064145
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr17_-_17947777 | 0.03 |
ENSRNOT00000036876
|
Rnf144b
|
ring finger protein 144B |
| chr8_+_122197027 | 0.03 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr15_-_24374753 | 0.03 |
ENSRNOT00000016274
|
Atg14
|
autophagy related 14 |
| chr3_+_100366168 | 0.03 |
ENSRNOT00000006689
|
Kif18a
|
kinesin family member 18A |
| chr12_-_14175945 | 0.03 |
ENSRNOT00000001469
|
Ap5z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chr1_+_220746387 | 0.03 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr3_-_148312791 | 0.03 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr2_-_196207560 | 0.03 |
ENSRNOT00000028589
|
Psmd4
|
proteasome 26S subunit, non-ATPase 4 |
| chrX_+_74205842 | 0.03 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr4_-_152380184 | 0.03 |
ENSRNOT00000091473
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr16_+_36116258 | 0.03 |
ENSRNOT00000017652
|
Sap30
|
Sin3A associated protein 30 |
| chr1_-_219450451 | 0.03 |
ENSRNOT00000025317
|
Rad9a
|
RAD9 checkpoint clamp component A |
| chr15_-_40545824 | 0.03 |
ENSRNOT00000017204
|
Nup58
|
nucleoporin 58 |
| chr8_-_48736506 | 0.03 |
ENSRNOT00000016227
|
Ccdc84
|
coiled-coil domain containing 84 |
| chr14_+_17064353 | 0.03 |
ENSRNOT00000003052
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr6_-_135829953 | 0.03 |
ENSRNOT00000080623
ENSRNOT00000039059 |
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr10_+_70118250 | 0.03 |
ENSRNOT00000009970
|
Zfp830
|
zinc finger protein 830 |
| chr4_-_118578815 | 0.02 |
ENSRNOT00000080502
|
Anxa4
|
annexin A4 |
| chr19_-_59384297 | 0.02 |
ENSRNOT00000077516
|
Tarbp1
|
TAR RNA binding protein 1 |
| chrX_-_105308554 | 0.02 |
ENSRNOT00000045556
|
Taf7l
|
TATA-box binding protein associated factor 7-like |
| chr3_-_124884570 | 0.02 |
ENSRNOT00000028887
|
Pcna
|
proliferating cell nuclear antigen |
| chr20_-_6864387 | 0.02 |
ENSRNOT00000068527
|
Ppil1
|
peptidylprolyl isomerase like 1 |
| chr1_-_213635546 | 0.02 |
ENSRNOT00000018861
|
Sirt3
|
sirtuin 3 |
| chr3_+_14990652 | 0.02 |
ENSRNOT00000090735
|
Dab2ip
|
DAB2 interacting protein |
| chr2_+_49682754 | 0.02 |
ENSRNOT00000079907
ENSRNOT00000085576 |
Emb
|
embigin |
| chr3_+_147422095 | 0.02 |
ENSRNOT00000006786
|
Angpt4
|
angiopoietin 4 |
| chr11_+_31806618 | 0.02 |
ENSRNOT00000043410
ENSRNOT00000002769 |
Son
|
Son DNA binding protein |
| chr3_+_151032952 | 0.02 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr10_-_2037891 | 0.02 |
ENSRNOT00000004563
|
Ercc4
|
ERCC excision repair 4, endonuclease catalytic subunit |
| chr15_-_37663584 | 0.02 |
ENSRNOT00000012093
|
Cryl1
|
crystallin, lambda 1 |
| chr3_+_175493698 | 0.02 |
ENSRNOT00000087356
|
Osbpl2
|
oxysterol binding protein-like 2 |
| chr5_-_74366817 | 0.02 |
ENSRNOT00000076705
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr4_-_51844331 | 0.02 |
ENSRNOT00000003593
|
Gpr37
|
G protein-coupled receptor 37 |
| chr15_+_2374582 | 0.02 |
ENSRNOT00000019644
|
Zfp503
|
zinc finger protein 503 |
| chr3_-_133131192 | 0.02 |
ENSRNOT00000055630
|
Tasp1
|
taspase 1 |
| chr5_-_138470096 | 0.02 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr18_+_24584900 | 0.02 |
ENSRNOT00000017075
|
Wdr33
|
WD repeat domain 33 |
| chr2_-_128675408 | 0.02 |
ENSRNOT00000019256
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr7_-_121302740 | 0.02 |
ENSRNOT00000067032
|
Rpl3
|
ribosomal protein L3 |
| chr12_-_43940798 | 0.02 |
ENSRNOT00000001485
|
RGD1562310
|
similar to hypothetical protein FLJ21415 |
| chr3_+_146695344 | 0.02 |
ENSRNOT00000010955
|
Gins1
|
GINS complex subunit 1 |
| chr4_+_77212203 | 0.02 |
ENSRNOT00000085805
|
Cul1
|
cullin 1 |
| chr8_+_128577345 | 0.02 |
ENSRNOT00000082356
|
Wdr48
|
WD repeat domain 48 |
| chr12_+_43940929 | 0.02 |
ENSRNOT00000001486
|
Rnft2
|
ring finger protein, transmembrane 2 |
| chr1_+_260153645 | 0.02 |
ENSRNOT00000054717
|
Zfp518a
|
zinc finger protein 518A |
| chr14_+_2613406 | 0.02 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr3_-_10161989 | 0.02 |
ENSRNOT00000012312
|
Exosc2
|
exosome component 2 |
| chr15_+_344685 | 0.02 |
ENSRNOT00000065542
ENSRNOT00000066928 |
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr10_+_975697 | 0.02 |
ENSRNOT00000089404
|
AC117889.1
|
|
| chr17_+_9837402 | 0.02 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr20_-_34929965 | 0.02 |
ENSRNOT00000004499
|
Mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr18_-_24693481 | 0.02 |
ENSRNOT00000021708
|
Sft2d3
|
SFT2 domain containing 3 |
| chr11_-_61748768 | 0.02 |
ENSRNOT00000078879
|
Ccdc191
|
coiled-coil domain containing 191 |
| chr13_+_56513286 | 0.02 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr5_+_155812105 | 0.02 |
ENSRNOT00000039341
|
AABR07073181.1
|
|
| chr12_+_13090172 | 0.02 |
ENSRNOT00000092558
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr8_-_36410612 | 0.02 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr8_-_23084879 | 0.02 |
ENSRNOT00000018874
|
Zfp653
|
zinc finger protein 653 |
| chr2_+_149214265 | 0.01 |
ENSRNOT00000084020
|
Med12l
|
mediator complex subunit 12-like |
| chr1_+_72380711 | 0.01 |
ENSRNOT00000022236
|
Fiz1
|
FLT3-interacting zinc finger 1 |
| chrX_+_27015884 | 0.01 |
ENSRNOT00000065814
|
Msl3
|
male-specific lethal 3 homolog (Drosophila) |
| chr9_+_15166118 | 0.01 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr3_-_175601127 | 0.01 |
ENSRNOT00000081226
|
Lama5
|
laminin subunit alpha 5 |
| chr19_+_55381565 | 0.01 |
ENSRNOT00000018923
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr5_+_157848206 | 0.01 |
ENSRNOT00000039663
ENSRNOT00000082138 |
Ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chrX_+_71540895 | 0.01 |
ENSRNOT00000004692
ENSRNOT00000082967 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr11_-_61748583 | 0.01 |
ENSRNOT00000093431
|
Ccdc191
|
coiled-coil domain containing 191 |
| chr1_+_174330695 | 0.01 |
ENSRNOT00000018497
|
Akip1
|
A-kinase interacting protein 1 |
| chr10_-_109979095 | 0.01 |
ENSRNOT00000073956
|
Dus1l
|
dihydrouridine synthase 1-like |
| chr11_+_61748883 | 0.01 |
ENSRNOT00000093552
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr1_+_154377247 | 0.01 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr18_+_3162543 | 0.01 |
ENSRNOT00000078615
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr8_+_118525682 | 0.01 |
ENSRNOT00000028288
|
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr10_-_71837851 | 0.01 |
ENSRNOT00000000258
|
Aatf
|
apoptosis antagonizing transcription factor |
| chr14_+_16491573 | 0.01 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr5_-_62001196 | 0.01 |
ENSRNOT00000012602
|
Trmo
|
tRNA methyltransferase O |
| chr5_-_152458023 | 0.01 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr6_+_788548 | 0.01 |
ENSRNOT00000005983
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 |
| chr8_+_36410683 | 0.01 |
ENSRNOT00000015177
|
Srpra
|
SRP receptor alpha subunit |
| chr9_+_65279675 | 0.01 |
ENSRNOT00000019486
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr8_-_21968415 | 0.01 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr8_+_128577080 | 0.01 |
ENSRNOT00000024172
|
Wdr48
|
WD repeat domain 48 |
| chr8_+_111210811 | 0.01 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr2_+_95008477 | 0.01 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chrX_+_71324365 | 0.01 |
ENSRNOT00000004911
|
Nono
|
non-POU domain containing, octamer-binding |
| chr7_-_117041693 | 0.01 |
ENSRNOT00000073835
|
Ccdc166
|
coiled-coil domain containing 166 |
| chr18_+_3163214 | 0.01 |
ENSRNOT00000017291
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr7_-_11353713 | 0.01 |
ENSRNOT00000061132
|
Zfr2
|
zinc finger RNA binding protein 2 |
| chr10_-_71441389 | 0.01 |
ENSRNOT00000003699
|
Tada2a
|
transcriptional adaptor 2A |
| chr2_-_225389120 | 0.01 |
ENSRNOT00000016739
|
Abcd3
|
ATP binding cassette subfamily D member 3 |
| chr8_+_22189600 | 0.01 |
ENSRNOT00000061100
|
Pde4a
|
phosphodiesterase 4A |
| chr7_-_2961873 | 0.01 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr7_-_31824064 | 0.01 |
ENSRNOT00000011494
ENSRNOT00000080824 |
Slc25a3
|
solute carrier family 25 member 3 |
| chr10_+_40553180 | 0.01 |
ENSRNOT00000087763
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr8_+_118378059 | 0.01 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr11_+_87220618 | 0.01 |
ENSRNOT00000041975
|
Gsc2
|
goosecoid homeobox 2 |
| chr10_-_67285617 | 0.01 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr9_-_73871888 | 0.01 |
ENSRNOT00000017686
|
Acadl
|
acyl-CoA dehydrogenase, long chain |
| chr6_+_110624856 | 0.01 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr2_-_30664163 | 0.01 |
ENSRNOT00000024801
|
Rad17
|
RAD17 checkpoint clamp loader component |
| chr4_-_177331874 | 0.01 |
ENSRNOT00000065387
ENSRNOT00000091099 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr10_+_80790168 | 0.01 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr12_+_51878153 | 0.01 |
ENSRNOT00000056798
|
Hscb
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr13_+_96111800 | 0.01 |
ENSRNOT00000005967
|
Desi2
|
desumoylating isopeptidase 2 |
| chr15_-_34338956 | 0.01 |
ENSRNOT00000026914
|
Mdp1
|
magnesium-dependent phosphatase 1 |
| chr4_-_129514390 | 0.01 |
ENSRNOT00000077530
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr19_+_58505811 | 0.01 |
ENSRNOT00000031717
|
Map10
|
microtubule-associated protein 10 |
| chr6_-_47811853 | 0.01 |
ENSRNOT00000010942
|
Allc
|
allantoicase |
| chr15_+_34297290 | 0.01 |
ENSRNOT00000081165
|
Rec8
|
REC8 meiotic recombination protein |
| chrX_-_63203643 | 0.01 |
ENSRNOT00000065194
ENSRNOT00000076974 |
Zfx
|
zinc finger protein X-linked |
| chr4_-_62438958 | 0.01 |
ENSRNOT00000014010
|
Wdr91
|
WD repeat domain 91 |
| chr19_-_10596851 | 0.01 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr7_+_141380322 | 0.01 |
ENSRNOT00000091207
|
Cox14
|
COX14, cytochrome c oxidase assembly factor |
| chr19_-_39246545 | 0.01 |
ENSRNOT00000036634
|
Pdf
|
peptide deformylase (mitochondrial) |
| chrX_-_35431164 | 0.01 |
ENSRNOT00000004968
|
Scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr2_+_182006242 | 0.01 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr1_-_84008293 | 0.01 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr19_-_55389256 | 0.01 |
ENSRNOT00000064180
|
Aprt
|
adenine phosphoribosyl transferase |
| chr4_-_147893992 | 0.01 |
ENSRNOT00000032158
|
Plxnd1
|
plexin D1 |
| chr17_-_32953641 | 0.01 |
ENSRNOT00000023332
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr1_+_80920747 | 0.01 |
ENSRNOT00000026077
|
Zfp180
|
zinc finger protein 180 |
| chr2_+_166403265 | 0.01 |
ENSRNOT00000012417
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr4_-_71227872 | 0.00 |
ENSRNOT00000050392
|
Tcaf2
|
TRPM8 channel-associated factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.0 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.0 | 0.0 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.0 | 0.0 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |