Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
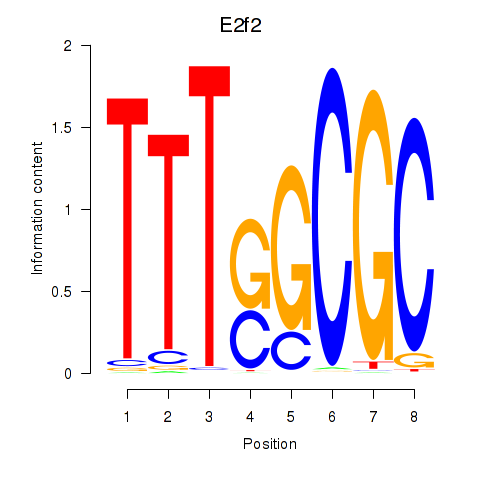
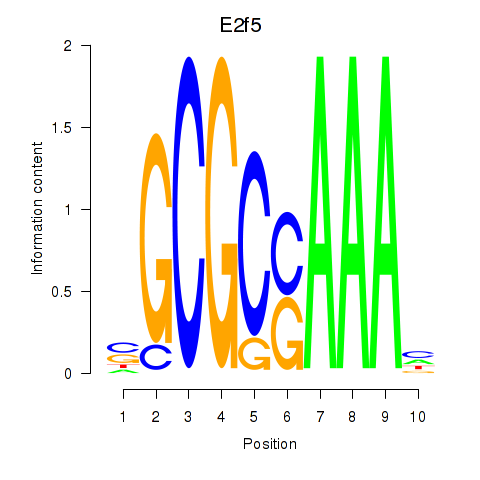
Results for E2f2_E2f5
Z-value: 0.29
Transcription factors associated with E2f2_E2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f2
|
ENSRNOG00000047741 | E2F transcription factor 2 |
|
E2f5
|
ENSRNOG00000010760 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f5 | rn6_v1_chr2_-_88366115_88366115 | -0.65 | 2.3e-01 | Click! |
| E2f2 | rn6_v1_chr5_+_154522119_154522119 | 0.40 | 5.0e-01 | Click! |
Activity profile of E2f2_E2f5 motif
Sorted Z-values of E2f2_E2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_42467586 | 0.14 |
ENSRNOT00000029416
|
Fbxo5
|
F-box protein 5 |
| chr8_+_117953444 | 0.11 |
ENSRNOT00000028141
|
Cdc25a
|
cell division cycle 25A |
| chr12_+_943006 | 0.10 |
ENSRNOT00000001449
|
Kl
|
Klotho |
| chr17_+_44520537 | 0.08 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr3_-_124884570 | 0.08 |
ENSRNOT00000028887
|
Pcna
|
proliferating cell nuclear antigen |
| chr6_+_144291974 | 0.07 |
ENSRNOT00000035255
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr3_+_125470551 | 0.07 |
ENSRNOT00000028898
|
Mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr12_-_17186679 | 0.07 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr4_+_81244469 | 0.06 |
ENSRNOT00000044407
ENSRNOT00000015828 |
Cbx3
|
chromobox 3 |
| chr10_+_72909550 | 0.06 |
ENSRNOT00000004540
|
Ppm1d
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr12_+_19314251 | 0.06 |
ENSRNOT00000001827
|
Ap4m1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr3_-_150073721 | 0.06 |
ENSRNOT00000022428
|
E2f1
|
E2F transcription factor 1 |
| chr6_-_91518996 | 0.05 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr12_-_19314016 | 0.05 |
ENSRNOT00000001825
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr8_+_132032944 | 0.05 |
ENSRNOT00000089278
|
Kif15
|
kinesin family member 15 |
| chr12_-_52452040 | 0.05 |
ENSRNOT00000067453
|
Pole
|
DNA polymerase epsilon, catalytic subunit |
| chr4_-_183426439 | 0.05 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chr10_-_108150511 | 0.05 |
ENSRNOT00000073337
|
Cbx8
|
chromobox 8 |
| chr2_+_243105674 | 0.05 |
ENSRNOT00000013919
|
H2afz
|
H2A histone family, member Z |
| chr8_+_12355767 | 0.05 |
ENSRNOT00000068445
|
Fam76b
|
family with sequence similarity 76, member B |
| chr4_-_120840111 | 0.05 |
ENSRNOT00000022231
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr5_+_144581427 | 0.05 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chr3_-_80052953 | 0.04 |
ENSRNOT00000018998
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr20_+_20576377 | 0.04 |
ENSRNOT00000000783
ENSRNOT00000086806 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr6_+_43884678 | 0.04 |
ENSRNOT00000091551
|
Rrm2
|
ribonucleotide reductase regulatory subunit M2 |
| chr8_+_108958046 | 0.04 |
ENSRNOT00000079618
ENSRNOT00000066917 |
Stag1
|
stromal antigen 1 |
| chr9_-_26932201 | 0.04 |
ENSRNOT00000017081
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr12_+_52452273 | 0.04 |
ENSRNOT00000056680
ENSRNOT00000088381 |
Pxmp2
|
peroxisomal membrane protein 2 |
| chr2_+_200187179 | 0.04 |
ENSRNOT00000093564
ENSRNOT00000025718 |
Notch2
|
notch 2 |
| chr7_+_97984862 | 0.04 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr19_+_14523554 | 0.04 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr8_-_110813000 | 0.04 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chrX_+_157353577 | 0.04 |
ENSRNOT00000090596
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr1_-_33275540 | 0.03 |
ENSRNOT00000017019
|
Irx2
|
iroquois homeobox 2 |
| chr5_+_47853818 | 0.03 |
ENSRNOT00000009228
ENSRNOT00000079656 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr3_-_125470413 | 0.03 |
ENSRNOT00000028893
|
Trmt6
|
tRNA methyltransferase 6 |
| chr4_-_77347011 | 0.03 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr17_+_9830332 | 0.03 |
ENSRNOT00000021946
|
Rab24
|
RAB24, member RAS oncogene family |
| chr13_+_104284660 | 0.03 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr10_-_13075864 | 0.03 |
ENSRNOT00000005220
|
Paqr4
|
progestin and adipoQ receptor family member 4 |
| chr9_-_93377643 | 0.03 |
ENSRNOT00000024712
|
Ncl
|
nucleolin |
| chr17_+_42302540 | 0.03 |
ENSRNOT00000025389
|
Gmnn
|
geminin, DNA replication inhibitor |
| chr6_+_126636662 | 0.03 |
ENSRNOT00000010739
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr17_-_88037034 | 0.03 |
ENSRNOT00000080193
ENSRNOT00000034907 |
Prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr4_-_157679962 | 0.03 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_-_44520240 | 0.02 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr8_-_68966108 | 0.02 |
ENSRNOT00000012155
|
Smad6
|
SMAD family member 6 |
| chr14_-_86706626 | 0.02 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr17_+_44763598 | 0.02 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr17_+_44528125 | 0.02 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr10_+_13061170 | 0.02 |
ENSRNOT00000004936
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr8_-_132032971 | 0.02 |
ENSRNOT00000005502
|
RGD1311745
|
similar to RIKEN cDNA 1110059G10 |
| chrX_+_157353730 | 0.02 |
ENSRNOT00000080691
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr9_+_9961021 | 0.02 |
ENSRNOT00000075767
|
Tubb4a
|
tubulin, beta 4A class IVa |
| chr4_-_145487426 | 0.02 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr15_+_2374582 | 0.02 |
ENSRNOT00000019644
|
Zfp503
|
zinc finger protein 503 |
| chr20_+_46429222 | 0.02 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr10_+_14373679 | 0.02 |
ENSRNOT00000022063
|
Ift140
|
intraflagellar transport 140 |
| chr1_+_203509731 | 0.02 |
ENSRNOT00000028007
|
Hmx3
|
H6 family homeobox 3 |
| chr14_-_79464770 | 0.02 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr1_-_89560719 | 0.02 |
ENSRNOT00000028653
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr5_-_16706909 | 0.02 |
ENSRNOT00000011314
|
Rps20
|
ribosomal protein S20 |
| chr5_+_114940053 | 0.02 |
ENSRNOT00000012396
|
Hook1
|
hook microtubule-tethering protein 1 |
| chr2_-_4195810 | 0.02 |
ENSRNOT00000018457
|
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr8_-_27852996 | 0.02 |
ENSRNOT00000037790
|
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr10_-_14324170 | 0.02 |
ENSRNOT00000035513
ENSRNOT00000090587 |
Hn1l
|
hematological and neurological expressed 1-like |
| chr2_-_26699333 | 0.02 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr1_-_201110928 | 0.02 |
ENSRNOT00000027721
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr3_-_113091770 | 0.02 |
ENSRNOT00000068132
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr7_-_94774569 | 0.02 |
ENSRNOT00000036399
|
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr1_+_264507985 | 0.02 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chr15_-_44627765 | 0.02 |
ENSRNOT00000058887
|
Dock5
|
dedicator of cytokinesis 5 |
| chr10_+_90622992 | 0.02 |
ENSRNOT00000032856
|
Meioc
|
meiosis specific with coiled-coil domain |
| chr11_-_27080701 | 0.02 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr14_-_20816588 | 0.02 |
ENSRNOT00000033806
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr1_+_174767960 | 0.02 |
ENSRNOT00000013362
|
Wee1
|
WEE1 G2 checkpoint kinase |
| chr6_+_11644578 | 0.02 |
ENSRNOT00000021923
ENSRNOT00000093689 |
Msh6
|
mutS homolog 6 |
| chr3_-_117766120 | 0.02 |
ENSRNOT00000056022
|
Fbn1
|
fibrillin 1 |
| chr13_+_110257571 | 0.02 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr3_+_110918243 | 0.02 |
ENSRNOT00000056432
|
Rad51
|
RAD51 recombinase |
| chr10_+_36098051 | 0.01 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr16_-_7103604 | 0.01 |
ENSRNOT00000033677
|
Gnl3
|
G protein nucleolar 3 |
| chr5_+_154668179 | 0.01 |
ENSRNOT00000015971
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr4_-_34282351 | 0.01 |
ENSRNOT00000011123
|
Rpa3
|
replication protein A3 |
| chr4_+_170820594 | 0.01 |
ENSRNOT00000032568
|
LOC500354
|
similar to C030030A07Rik protein |
| chr2_-_181874223 | 0.01 |
ENSRNOT00000035846
|
Rbm46
|
RNA binding motif protein 46 |
| chr17_+_11953552 | 0.01 |
ENSRNOT00000090782
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr6_+_108167716 | 0.01 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr1_+_167538744 | 0.01 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr2_+_128461224 | 0.01 |
ENSRNOT00000018872
|
Jade1
|
jade family PHD finger 1 |
| chr8_-_82351108 | 0.01 |
ENSRNOT00000013053
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr17_+_17965881 | 0.01 |
ENSRNOT00000021751
|
Dek
|
DEK proto-oncogene |
| chr18_-_33414236 | 0.01 |
ENSRNOT00000020385
|
Yipf5
|
Yip1 domain family, member 5 |
| chr17_+_77224112 | 0.01 |
ENSRNOT00000024178
|
Mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr9_+_17949764 | 0.01 |
ENSRNOT00000027264
|
Cdc5l
|
cell division cycle 5-like |
| chr3_+_2158995 | 0.01 |
ENSRNOT00000010270
|
Zmynd19
|
zinc finger, MYND-type containing 19 |
| chr5_+_152721940 | 0.01 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr8_-_65587658 | 0.01 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_+_101884019 | 0.01 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chr13_+_73196571 | 0.01 |
ENSRNOT00000004772
|
Acbd6
|
acyl-CoA binding domain containing 6 |
| chr5_-_58455819 | 0.01 |
ENSRNOT00000078082
|
Fancg
|
Fanconi anemia, complementation group G |
| chr15_+_3936786 | 0.01 |
ENSRNOT00000066163
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr17_+_16333415 | 0.01 |
ENSRNOT00000060550
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_154722220 | 0.01 |
ENSRNOT00000089743
ENSRNOT00000087893 |
Fmr1
|
fragile X mental retardation 1 |
| chr1_+_142034423 | 0.01 |
ENSRNOT00000090006
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr6_+_126636491 | 0.01 |
ENSRNOT00000089601
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr8_-_97494834 | 0.01 |
ENSRNOT00000080421
|
Morf4l1
|
mortality factor 4 like 1 |
| chr13_+_48679774 | 0.01 |
ENSRNOT00000075674
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr1_+_198214797 | 0.01 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr1_-_198706852 | 0.01 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr3_+_146695344 | 0.01 |
ENSRNOT00000010955
|
Gins1
|
GINS complex subunit 1 |
| chr8_-_65587427 | 0.01 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr19_-_37245217 | 0.01 |
ENSRNOT00000020927
|
MGC116202
|
hypothetical protein LOC688735 |
| chr2_+_207108552 | 0.01 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr17_-_1672021 | 0.01 |
ENSRNOT00000036287
|
Zfp367
|
zinc finger protein 367 |
| chr8_+_127789048 | 0.01 |
ENSRNOT00000052275
|
Dlec1
|
deleted in lung and esophageal cancer 1 |
| chr17_+_44738643 | 0.01 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr15_-_88622413 | 0.01 |
ENSRNOT00000092174
|
Pou4f1
|
POU class 4 homeobox 1 |
| chr3_-_59688692 | 0.01 |
ENSRNOT00000078752
|
Sp3
|
Sp3 transcription factor |
| chr7_+_11784341 | 0.01 |
ENSRNOT00000026155
|
Plekhj1
|
pleckstrin homology domain containing J1 |
| chr10_-_6870011 | 0.01 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr20_+_5114013 | 0.01 |
ENSRNOT00000076738
|
Gpank1
|
G patch domain and ankyrin repeats 1 |
| chr20_+_6049286 | 0.01 |
ENSRNOT00000042143
ENSRNOT00000092566 |
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr3_-_164964702 | 0.01 |
ENSRNOT00000014595
|
Adnp
|
activity-dependent neuroprotector homeobox |
| chr1_+_221448661 | 0.01 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chr19_+_25262076 | 0.01 |
ENSRNOT00000030658
|
RGD1306072
|
hypothetical LOC304654 |
| chr17_+_43734461 | 0.01 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr5_-_173659675 | 0.01 |
ENSRNOT00000027547
|
Klhl17
|
kelch-like family member 17 |
| chr16_-_38063821 | 0.01 |
ENSRNOT00000077452
ENSRNOT00000089533 |
Glra3
|
glycine receptor, alpha 3 |
| chr1_-_21854763 | 0.01 |
ENSRNOT00000089196
ENSRNOT00000020528 |
Ctgf
|
connective tissue growth factor |
| chr9_+_93545396 | 0.01 |
ENSRNOT00000025093
|
LOC100359583
|
hypothetical protein LOC100359583 |
| chr1_+_228337767 | 0.01 |
ENSRNOT00000066247
|
Patl1
|
PAT1 homolog 1, processing body mRNA decay factor |
| chrX_+_54734385 | 0.01 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr5_-_128186532 | 0.01 |
ENSRNOT00000012635
|
Prpf38a
|
pre-mRNA processing factor 38A |
| chrX_+_40363646 | 0.01 |
ENSRNOT00000010135
|
Sms
|
spermine synthase |
| chr10_+_67427066 | 0.01 |
ENSRNOT00000035642
|
Atad5
|
ATPase family, AAA domain containing 5 |
| chr19_-_9843673 | 0.01 |
ENSRNOT00000015795
|
Gins3
|
GINS complex subunit 3 |
| chr17_-_44748188 | 0.01 |
ENSRNOT00000081970
|
LOC103690190
|
histone H2A type 1-E |
| chr16_+_7103998 | 0.01 |
ENSRNOT00000016581
|
Pbrm1
|
polybromo 1 |
| chr13_-_80703615 | 0.01 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr15_+_28377996 | 0.01 |
ENSRNOT00000078475
|
Arhgef40
|
Rho guanine nucleotide exchange factor 40 |
| chr2_+_190007216 | 0.01 |
ENSRNOT00000015612
|
S100a6
|
S100 calcium binding protein A6 |
| chr17_+_8299131 | 0.01 |
ENSRNOT00000083687
|
Gm45623
|
predicted gene 45623 |
| chr16_-_37908173 | 0.01 |
ENSRNOT00000075026
|
Glra3
|
glycine receptor, alpha 3 |
| chr10_-_46206135 | 0.01 |
ENSRNOT00000091471
|
Cops3
|
COP9 signalosome subunit 3 |
| chr15_-_83442008 | 0.01 |
ENSRNOT00000039400
|
Mzt1
|
mitotic spindle organizing protein 1 |
| chr1_+_273563441 | 0.01 |
ENSRNOT00000048691
|
RGD1561333
|
similar to 60S ribosomal protein L8 |
| chr2_-_84608712 | 0.01 |
ENSRNOT00000028561
|
March6
|
membrane associated ring-CH-type finger 6 |
| chr6_-_126636484 | 0.01 |
ENSRNOT00000088000
ENSRNOT00000063786 |
Gon7
|
GON7, KEOPS complex subunit homolog |
| chr1_+_48033768 | 0.01 |
ENSRNOT00000020064
|
Mrpl18
|
mitochondrial ribosomal protein L18 |
| chr16_+_80729400 | 0.01 |
ENSRNOT00000036383
|
Tdrp
|
testis development related protein |
| chr7_-_49741540 | 0.00 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr16_+_80729959 | 0.00 |
ENSRNOT00000082049
|
Tdrp
|
testis development related protein |
| chr10_-_62699723 | 0.00 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr6_-_103935122 | 0.00 |
ENSRNOT00000058354
|
LOC500684
|
hypothetical protein LOC500684 |
| chr1_-_31262466 | 0.00 |
ENSRNOT00000029531
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr7_+_27438351 | 0.00 |
ENSRNOT00000077976
|
AABR07056503.1
|
|
| chr4_+_62299044 | 0.00 |
ENSRNOT00000032077
|
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr11_-_89510871 | 0.00 |
ENSRNOT00000035247
|
Prkdc
|
protein kinase, DNA activated, catalytic polypeptide |
| chr10_-_36716601 | 0.00 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr7_-_136853154 | 0.00 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr14_-_44375804 | 0.00 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr6_-_2886465 | 0.00 |
ENSRNOT00000038448
|
Srsf7
|
serine and arginine rich splicing factor 7 |
| chr1_+_167538263 | 0.00 |
ENSRNOT00000074058
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr19_-_52206310 | 0.00 |
ENSRNOT00000087886
|
Mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr10_+_11206226 | 0.00 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr1_-_250727079 | 0.00 |
ENSRNOT00000079942
ENSRNOT00000087588 |
Sgms1
|
sphingomyelin synthase 1 |
| chr15_+_4356261 | 0.00 |
ENSRNOT00000009009
|
Dnajc9
|
DnaJ heat shock protein family (Hsp40) member C9 |
| chr15_+_83442144 | 0.00 |
ENSRNOT00000039344
|
Bora
|
bora, aurora kinase A activator |
| chrX_+_113510621 | 0.00 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr3_+_111597102 | 0.00 |
ENSRNOT00000081462
|
Tyro3
|
TYRO3 protein tyrosine kinase |
| chrX_-_128268285 | 0.00 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr15_+_45422010 | 0.00 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr1_-_80630038 | 0.00 |
ENSRNOT00000025281
|
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr10_-_59743315 | 0.00 |
ENSRNOT00000093646
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr10_+_105796680 | 0.00 |
ENSRNOT00000000264
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr1_-_218810118 | 0.00 |
ENSRNOT00000065950
ENSRNOT00000020886 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr9_+_81672758 | 0.00 |
ENSRNOT00000020646
|
Ctdsp1
|
CTD small phosphatase 1 |
| chr10_+_4313100 | 0.00 |
ENSRNOT00000074487
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr10_-_74119009 | 0.00 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr14_-_35149608 | 0.00 |
ENSRNOT00000003050
ENSRNOT00000090654 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr6_+_132805794 | 0.00 |
ENSRNOT00000006281
|
Wdr25
|
WD repeat domain 25 |
| chr7_+_122636171 | 0.00 |
ENSRNOT00000068666
|
Xpnpep3
|
X-prolyl aminopeptidase 3 |
| chr13_-_101697684 | 0.00 |
ENSRNOT00000078834
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr1_-_82546937 | 0.00 |
ENSRNOT00000028100
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr12_+_660011 | 0.00 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr13_-_45068077 | 0.00 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr3_-_176431423 | 0.00 |
ENSRNOT00000037271
|
Ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr9_-_10293980 | 0.00 |
ENSRNOT00000072703
|
Dus3l
|
dihydrouridine synthase 3-like |
| chr11_+_89511191 | 0.00 |
ENSRNOT00000002510
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr8_+_71514281 | 0.00 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr15_+_61731735 | 0.00 |
ENSRNOT00000015517
|
Kbtbd6
|
kelch repeat and BTB domain containing 6 |
| chr1_+_264670841 | 0.00 |
ENSRNOT00000034814
ENSRNOT00000082412 |
Slf2
|
SMC5-SMC6 complex localization factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f2_E2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |