Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
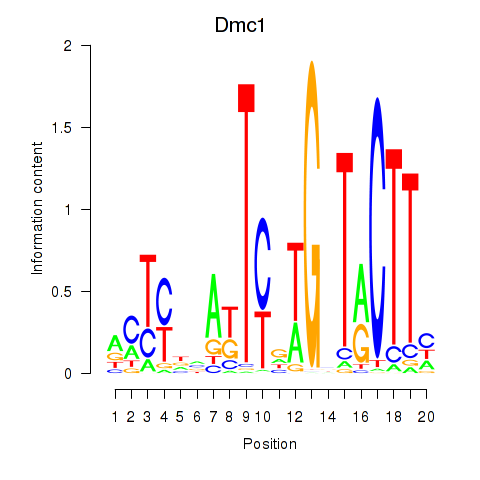
Results for Dmc1
Z-value: 0.91
Transcription factors associated with Dmc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dmc1
|
ENSRNOG00000013807 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmc1 | rn6_v1_chr7_-_120839577_120839577 | 0.20 | 7.5e-01 | Click! |
Activity profile of Dmc1 motif
Sorted Z-values of Dmc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_14599594 | 0.36 |
ENSRNOT00000018138
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr6_-_87427153 | 0.34 |
ENSRNOT00000071999
|
AABR07064622.1
|
|
| chr11_+_82052774 | 0.34 |
ENSRNOT00000062045
|
AABR07034637.1
|
|
| chr8_-_104155775 | 0.32 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr18_+_25613831 | 0.30 |
ENSRNOT00000091040
|
Tslp
|
thymic stromal lymphopoietin |
| chr10_-_38969501 | 0.30 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr15_+_31141142 | 0.29 |
ENSRNOT00000060304
|
LOC103693854
|
uncharacterized LOC103693854 |
| chr4_-_117584819 | 0.29 |
ENSRNOT00000090815
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chrX_+_137934484 | 0.28 |
ENSRNOT00000049046
|
LOC317588
|
hypothetical protein LOC317588 |
| chr2_-_208623314 | 0.28 |
ENSRNOT00000022731
|
Pifo
|
primary cilia formation |
| chr15_+_32051651 | 0.27 |
ENSRNOT00000071887
|
LOC103693854
|
uncharacterized LOC103693854 |
| chr5_-_74029238 | 0.26 |
ENSRNOT00000031432
|
Frrs1l
|
ferric-chelate reductase 1-like |
| chr4_+_170347410 | 0.26 |
ENSRNOT00000040508
|
LOC500350
|
LRRGT00139 |
| chr10_+_105630810 | 0.25 |
ENSRNOT00000064904
|
Prcd
|
photoreceptor disc component |
| chrX_+_115208863 | 0.24 |
ENSRNOT00000072094
|
AABR07040936.1
|
|
| chr4_+_22859622 | 0.24 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr1_+_65576535 | 0.23 |
ENSRNOT00000026575
|
Slc27a5
|
solute carrier family 27 member 5 |
| chr10_-_88152064 | 0.23 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chr4_+_102147211 | 0.23 |
ENSRNOT00000083239
|
AABR07060980.1
|
|
| chr16_+_22361998 | 0.22 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr10_-_94500591 | 0.21 |
ENSRNOT00000015976
|
Cd79b
|
CD79b molecule |
| chr8_-_84522588 | 0.21 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr7_+_20262680 | 0.21 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr7_-_94774569 | 0.21 |
ENSRNOT00000036399
|
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr12_-_17322608 | 0.20 |
ENSRNOT00000033038
|
LOC102546864
|
uncharacterized LOC102546864 |
| chr11_-_64952687 | 0.20 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr7_+_1720160 | 0.20 |
ENSRNOT00000073169
|
AABR07055353.1
|
|
| chr8_-_119889661 | 0.20 |
ENSRNOT00000011780
|
Stac
|
SH3 and cysteine rich domain |
| chr2_-_23256158 | 0.19 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr2_-_47281421 | 0.19 |
ENSRNOT00000086114
|
Itga1
|
integrin subunit alpha 1 |
| chr19_+_54766589 | 0.19 |
ENSRNOT00000025894
|
Banp
|
Btg3 associated nuclear protein |
| chr1_+_201687758 | 0.19 |
ENSRNOT00000093308
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr2_+_149899836 | 0.19 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr2_+_180012414 | 0.19 |
ENSRNOT00000082273
|
Pdgfc
|
platelet derived growth factor C |
| chr13_-_113817995 | 0.19 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chr7_+_141642777 | 0.17 |
ENSRNOT00000079811
|
AABR07058884.2
|
|
| chr6_+_52702544 | 0.17 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr4_+_10295122 | 0.17 |
ENSRNOT00000017374
|
Ccdc146
|
coiled-coil domain containing 146 |
| chr8_+_52729003 | 0.17 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr16_-_49820235 | 0.17 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_+_38727048 | 0.16 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr12_-_5490935 | 0.16 |
ENSRNOT00000050885
|
Zfp958
|
zinc finger protein 958 |
| chr10_+_88620655 | 0.16 |
ENSRNOT00000055248
|
Hspb9
|
heat shock protein family B (small) member 9 |
| chr17_+_43868191 | 0.16 |
ENSRNOT00000059403
|
Btn2a2
|
butyrophilin, subfamily 2, member A2 |
| chr17_+_45175121 | 0.16 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr3_+_46185360 | 0.16 |
ENSRNOT00000051344
|
LOC100361645
|
LRRGT00075-like |
| chr6_-_50941248 | 0.16 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr13_+_85818427 | 0.15 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr1_-_225631468 | 0.15 |
ENSRNOT00000072579
|
AABR07006232.1
|
|
| chr15_+_4077951 | 0.15 |
ENSRNOT00000085266
|
Myoz1
|
myozenin 1 |
| chr9_+_72933304 | 0.15 |
ENSRNOT00000041632
|
Crygf
|
crystallin, gamma F |
| chr15_-_93748742 | 0.15 |
ENSRNOT00000093370
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr2_+_242882306 | 0.15 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr7_-_68549763 | 0.15 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chrX_-_25628272 | 0.14 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr4_-_117767772 | 0.14 |
ENSRNOT00000084170
|
LOC103690120
|
probable N-acetyltransferase CML1 |
| chr8_-_55696601 | 0.14 |
ENSRNOT00000016127
|
RGD1562914
|
RGD1562914 |
| chr1_+_142679345 | 0.14 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr1_-_24858316 | 0.14 |
ENSRNOT00000077927
|
AABR07000733.1
|
|
| chr1_+_48077033 | 0.14 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr4_-_119591817 | 0.14 |
ENSRNOT00000048768
|
AC097129.1
|
|
| chr11_+_82848853 | 0.14 |
ENSRNOT00000073743
|
Thpo
|
thrombopoietin |
| chr1_-_247486202 | 0.14 |
ENSRNOT00000088193
|
Rln1
|
relaxin 1 |
| chr1_+_248647170 | 0.14 |
ENSRNOT00000015016
|
Tpd52l3
|
tumor protein D52-like 3 |
| chr4_+_183896303 | 0.14 |
ENSRNOT00000055437
|
RGD1309621
|
similar to hypothetical protein FLJ10652 |
| chr9_+_94178221 | 0.14 |
ENSRNOT00000033487
|
Alppl2
|
alkaline phosphatase, placental-like 2 |
| chr9_+_16924520 | 0.14 |
ENSRNOT00000025094
|
Slc22a7
|
solute carrier family 22 member 7 |
| chr15_-_33656089 | 0.14 |
ENSRNOT00000024186
|
Myh7
|
myosin heavy chain 7 |
| chr8_+_55196758 | 0.13 |
ENSRNOT00000065879
|
Fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr5_+_144106802 | 0.13 |
ENSRNOT00000035637
ENSRNOT00000079779 |
Lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr2_-_185303610 | 0.13 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr4_-_161091010 | 0.13 |
ENSRNOT00000043708
|
Senp17
|
Sumo1/sentrin/SMT3 specific peptidase 17 |
| chr12_+_4737817 | 0.13 |
ENSRNOT00000035992
|
AABR07035107.1
|
|
| chr8_+_44136496 | 0.13 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr19_-_29134876 | 0.13 |
ENSRNOT00000040545
|
AABR07043523.1
|
|
| chrM_+_9870 | 0.13 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr3_-_162059524 | 0.13 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr16_-_21473808 | 0.13 |
ENSRNOT00000066776
|
Zfp964
|
zinc finger protein 964 |
| chr13_+_89386023 | 0.13 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr2_+_204932159 | 0.13 |
ENSRNOT00000078376
|
Ngf
|
nerve growth factor |
| chr19_+_26416818 | 0.13 |
ENSRNOT00000040473
|
AABR07043200.1
|
|
| chr10_+_65552897 | 0.13 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr1_+_65851060 | 0.13 |
ENSRNOT00000036880
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr16_-_6404578 | 0.12 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr1_-_157461588 | 0.12 |
ENSRNOT00000068402
|
Ankrd42
|
ankyrin repeat domain 42 |
| chr8_+_29714285 | 0.12 |
ENSRNOT00000042890
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr15_+_62406873 | 0.12 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr1_-_167698263 | 0.12 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr2_-_154508641 | 0.12 |
ENSRNOT00000065346
|
RGD1565059
|
similar to hypothetical protein E130311K13 |
| chr5_+_6373583 | 0.12 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr4_-_176026133 | 0.12 |
ENSRNOT00000043374
ENSRNOT00000046598 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr2_-_209973205 | 0.12 |
ENSRNOT00000024593
|
Cym
|
chymosin |
| chr13_+_51958834 | 0.12 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr1_+_189233141 | 0.12 |
ENSRNOT00000049380
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr5_-_127661028 | 0.12 |
ENSRNOT00000015466
|
Podn
|
podocan |
| chr3_-_46726946 | 0.11 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr1_-_228753422 | 0.11 |
ENSRNOT00000028626
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr14_+_58877806 | 0.11 |
ENSRNOT00000051559
|
RGD1562755
|
similar to 60S ribosomal protein L23a |
| chr8_+_82288705 | 0.11 |
ENSRNOT00000012409
|
Bcl2l10
|
BCL2 like 10 |
| chr5_-_133959447 | 0.11 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr4_-_85314672 | 0.11 |
ENSRNOT00000033672
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr4_-_119327822 | 0.11 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr5_-_68059933 | 0.11 |
ENSRNOT00000088716
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr13_-_110257367 | 0.11 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr2_+_20857202 | 0.11 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr1_-_167884690 | 0.11 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr7_-_18577325 | 0.11 |
ENSRNOT00000084308
ENSRNOT00000090849 |
March2
|
membrane associated ring-CH-type finger 2 |
| chr3_-_161040511 | 0.11 |
ENSRNOT00000037518
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr1_-_166919302 | 0.11 |
ENSRNOT00000026925
|
Folr2
|
folate receptor beta |
| chr4_-_78458179 | 0.11 |
ENSRNOT00000078473
ENSRNOT00000011327 |
Tmem176b
|
transmembrane protein 176B |
| chr12_+_45319501 | 0.11 |
ENSRNOT00000090630
ENSRNOT00000041732 |
RGD1561114
|
similar to hypothetical protein 4930474N05 |
| chrM_+_9451 | 0.10 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_+_85427555 | 0.10 |
ENSRNOT00000015469
|
Fam188b
|
family with sequence similarity 188, member B |
| chr2_+_252090669 | 0.10 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr1_+_48319522 | 0.10 |
ENSRNOT00000080281
|
AC114389.1
|
|
| chr5_+_154800226 | 0.10 |
ENSRNOT00000016046
|
Htr1d
|
5-hydroxytryptamine receptor 1D |
| chr1_-_239265997 | 0.10 |
ENSRNOT00000037500
|
RGD1359158
|
similar to RIKEN cDNA 1110059E24 |
| chr17_-_21677477 | 0.10 |
ENSRNOT00000035448
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr1_+_73837944 | 0.10 |
ENSRNOT00000036413
|
Lair1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr15_+_41799992 | 0.10 |
ENSRNOT00000032631
|
AABR07072463.1
|
|
| chr2_+_104290726 | 0.10 |
ENSRNOT00000017387
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr1_+_256382791 | 0.10 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr4_+_150199627 | 0.10 |
ENSRNOT00000052259
|
AABR07061871.1
|
|
| chr7_-_140417530 | 0.10 |
ENSRNOT00000077884
|
Fkbp11
|
FK506 binding protein 11 |
| chr15_-_59215803 | 0.10 |
ENSRNOT00000032301
|
Lacc1
|
laccase domain containing 1 |
| chr6_+_64224861 | 0.10 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr4_-_100883038 | 0.10 |
ENSRNOT00000041880
|
LOC100364435
|
thymosin, beta 10-like |
| chr7_+_48867664 | 0.10 |
ENSRNOT00000005862
|
Ppfia2
|
PTPRF interacting protein alpha 2 |
| chr1_-_260254600 | 0.10 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr16_+_40050734 | 0.10 |
ENSRNOT00000067375
|
Spcs3
|
signal peptidase complex subunit 3 |
| chr1_-_197858016 | 0.10 |
ENSRNOT00000074778
|
Atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr4_-_6046477 | 0.10 |
ENSRNOT00000073925
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr10_-_70802782 | 0.10 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr11_+_67101714 | 0.10 |
ENSRNOT00000078936
|
Cd86
|
CD86 molecule |
| chr8_-_49502647 | 0.10 |
ENSRNOT00000040313
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr18_-_7081356 | 0.10 |
ENSRNOT00000021253
|
Chst9
|
carbohydrate sulfotransferase 9 |
| chr18_+_55505993 | 0.10 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr3_-_153468221 | 0.10 |
ENSRNOT00000010443
|
Ghrh
|
growth hormone releasing hormone |
| chr3_-_161050557 | 0.10 |
ENSRNOT00000019829
|
Eppin
|
epididymal peptidase inhibitor |
| chr4_-_170740274 | 0.10 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr8_+_23492621 | 0.09 |
ENSRNOT00000093071
|
Bbs9
|
Bardet-Biedl syndrome 9 |
| chrX_+_14019961 | 0.09 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr6_-_76079664 | 0.09 |
ENSRNOT00000033533
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr13_+_52976507 | 0.09 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr10_-_49196177 | 0.09 |
ENSRNOT00000084418
|
Zfp286a
|
zinc finger protein 286A |
| chr15_+_28319136 | 0.09 |
ENSRNOT00000048723
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr9_+_111220858 | 0.09 |
ENSRNOT00000076669
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr4_-_64831473 | 0.09 |
ENSRNOT00000033268
|
Dgki
|
diacylglycerol kinase, iota |
| chr1_+_265298868 | 0.09 |
ENSRNOT00000023278
|
Dpcd
|
deleted in primary ciliary dyskinesia |
| chrM_-_14061 | 0.09 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr9_+_12114977 | 0.09 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr7_-_117732339 | 0.09 |
ENSRNOT00000092917
ENSRNOT00000020696 |
Foxh1
|
forkhead box H1 |
| chr8_-_27852996 | 0.09 |
ENSRNOT00000037790
|
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr1_+_215609036 | 0.09 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr11_-_70618347 | 0.09 |
ENSRNOT00000002435
|
Zfp148
|
zinc finger protein 148 |
| chr10_+_45893018 | 0.09 |
ENSRNOT00000004280
ENSRNOT00000086710 |
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr13_-_51784639 | 0.09 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr13_-_90832469 | 0.09 |
ENSRNOT00000086508
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr7_-_3246071 | 0.09 |
ENSRNOT00000044292
|
Ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr2_-_157035483 | 0.09 |
ENSRNOT00000076491
|
LOC304239
|
similar to RalA binding protein 1 |
| chr14_+_17210733 | 0.09 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr3_+_138174054 | 0.09 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr5_-_152589719 | 0.09 |
ENSRNOT00000022522
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr6_+_126040631 | 0.09 |
ENSRNOT00000008886
|
Slc24a4
|
solute carrier family 24 member 4 |
| chr1_-_101360971 | 0.09 |
ENSRNOT00000028164
|
Lin7b
|
lin-7 homolog B, crumbs cell polarity complex component |
| chr3_-_110828879 | 0.09 |
ENSRNOT00000013963
|
Ccdc32
|
coiled-coil domain containing 32 |
| chr1_+_190462327 | 0.09 |
ENSRNOT00000030732
|
LOC691519
|
similar to ankyrin repeat domain 26 |
| chr16_-_3762877 | 0.09 |
ENSRNOT00000052224
|
Duxbl1
|
double homeobox B-like 1 |
| chr10_-_10725655 | 0.09 |
ENSRNOT00000061236
|
Ubn1
|
ubinuclein 1 |
| chr10_-_36419926 | 0.09 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr1_-_88387424 | 0.09 |
ENSRNOT00000089393
|
LOC102547241
|
zinc finger protein 82-like |
| chr15_+_4850122 | 0.09 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr14_+_23611735 | 0.09 |
ENSRNOT00000031074
|
Cenpc
|
centromere protein C |
| chr16_-_81797815 | 0.09 |
ENSRNOT00000026666
|
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr12_+_12649861 | 0.09 |
ENSRNOT00000092402
|
Ccz1b
|
CCZ1 homolog B, vacuolar protein trafficking and biogenesis associated |
| chr17_+_49417067 | 0.09 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr1_-_88908750 | 0.09 |
ENSRNOT00000028297
|
Aplp1
|
amyloid beta precursor like protein 1 |
| chr3_-_105214989 | 0.09 |
ENSRNOT00000037895
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr8_+_48569328 | 0.09 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_-_103323476 | 0.08 |
ENSRNOT00000019051
|
Mrgprx3
|
MAS related GPR family member X3 |
| chr5_+_47546014 | 0.08 |
ENSRNOT00000065882
|
Bach2
|
BTB domain and CNC homolog 2 |
| chr7_+_28066635 | 0.08 |
ENSRNOT00000005844
|
Pah
|
phenylalanine hydroxylase |
| chrX_+_120859968 | 0.08 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr5_+_3675833 | 0.08 |
ENSRNOT00000011083
|
RGD1564405
|
similar to mKIAA1321 protein |
| chr11_+_88095170 | 0.08 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr1_-_140835151 | 0.08 |
ENSRNOT00000032174
|
Hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr15_+_30612409 | 0.08 |
ENSRNOT00000072977
|
AABR07017748.1
|
|
| chr20_-_6257604 | 0.08 |
ENSRNOT00000092489
|
Stk38
|
serine/threonine kinase 38 |
| chr19_+_55246926 | 0.08 |
ENSRNOT00000017358
|
Il17c
|
interleukin 17C |
| chr5_+_60850852 | 0.08 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
| chr2_-_62034628 | 0.08 |
ENSRNOT00000074446
|
Sub1
|
SUB1 homolog, transcriptional regulator |
| chr19_+_41968705 | 0.08 |
ENSRNOT00000020365
|
Ist1
|
IST1, ESCRT-III associated factor |
| chr5_-_113939127 | 0.08 |
ENSRNOT00000039554
|
Mysm1
|
myb-like, SWIRM and MPN domains 1 |
| chr8_+_48571323 | 0.08 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_-_205750786 | 0.08 |
ENSRNOT00000023946
|
Mmp21
|
matrix metallopeptidase 21 |
| chr14_+_17534412 | 0.08 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chrX_+_984798 | 0.08 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr16_-_61791091 | 0.08 |
ENSRNOT00000042003
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr4_-_150616895 | 0.08 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr19_+_39067363 | 0.08 |
ENSRNOT00000083515
|
Has3
|
hyaluronan synthase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dmc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 0.3 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.1 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.2 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0003163 | sinoatrial node development(GO:0003163) |
| 0.0 | 0.1 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.1 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0060060 | photoreceptor cell morphogenesis(GO:0008594) post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.0 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.0 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.0 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.0 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:2000872 | positive regulation of female gonad development(GO:2000196) positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.0 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0019660 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.0 | 0.2 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.0 | 0.1 | GO:0071209 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0004321 | acyl-CoA ligase activity(GO:0003996) fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |