Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
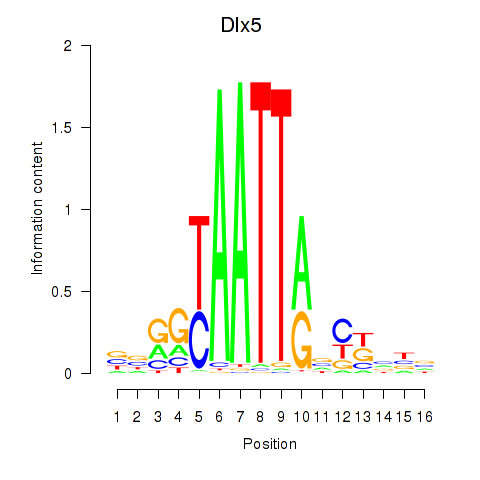
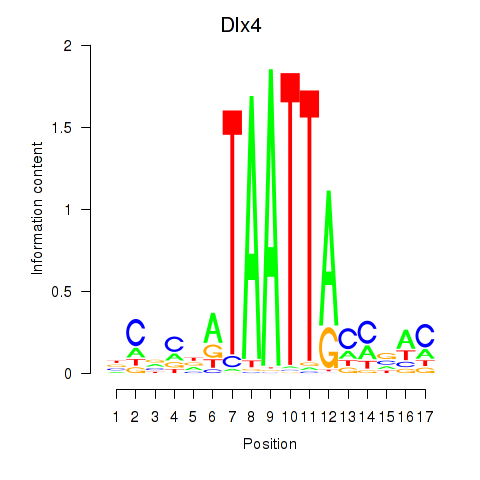
Results for Dlx5_Dlx4
Z-value: 0.55
Transcription factors associated with Dlx5_Dlx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dlx5
|
ENSRNOG00000010905 | distal-less homeobox 5 |
|
Dlx4
|
ENSRNOG00000004399 | distal-less homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx5 | rn6_v1_chr4_-_32392007_32392007 | -0.55 | 3.3e-01 | Click! |
| Dlx4 | rn6_v1_chr10_-_82963919_82963919 | -0.15 | 8.1e-01 | Click! |
Activity profile of Dlx5_Dlx4 motif
Sorted Z-values of Dlx5_Dlx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_66417741 | 0.60 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr2_+_145174876 | 0.55 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr2_-_185852759 | 0.41 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr20_-_9855443 | 0.30 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr7_-_13751271 | 0.28 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chr12_-_17186679 | 0.27 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr5_-_151459037 | 0.26 |
ENSRNOT00000064472
ENSRNOT00000087836 |
Sytl1
|
synaptotagmin-like 1 |
| chrM_+_7758 | 0.23 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr4_+_148782479 | 0.22 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr2_+_58724855 | 0.21 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr4_+_94696965 | 0.20 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chrX_-_123980357 | 0.18 |
ENSRNOT00000049435
|
Rhox8
|
reproductive homeobox 8 |
| chr1_-_71373605 | 0.17 |
ENSRNOT00000034854
|
Galp
|
galanin-like peptide |
| chrM_+_9451 | 0.17 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr7_-_139223116 | 0.16 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr15_+_87722221 | 0.16 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr5_-_22769907 | 0.15 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr17_-_2705123 | 0.15 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr17_+_24416651 | 0.14 |
ENSRNOT00000024458
|
Cd83
|
CD83 molecule |
| chr2_-_147819335 | 0.14 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr10_-_63176463 | 0.14 |
ENSRNOT00000004717
|
Slc6a4
|
solute carrier family 6 member 4 |
| chr6_-_123577695 | 0.14 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chrM_+_3904 | 0.13 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_-_38538987 | 0.13 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr7_+_144052061 | 0.13 |
ENSRNOT00000020103
ENSRNOT00000091378 |
Amhr2
|
anti-Mullerian hormone receptor type 2 |
| chr5_+_6373583 | 0.12 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr9_+_95161157 | 0.12 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr5_+_24410863 | 0.12 |
ENSRNOT00000010591
|
Tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr2_+_187951344 | 0.11 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chrX_+_131617798 | 0.11 |
ENSRNOT00000074384
|
Prr32
|
proline rich 32 |
| chr4_+_157726941 | 0.11 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr2_+_178117466 | 0.11 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr3_-_7498555 | 0.11 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr10_-_88670430 | 0.10 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr1_-_198104109 | 0.10 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr1_+_264260505 | 0.10 |
ENSRNOT00000018815
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr2_-_205212681 | 0.10 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr7_-_54855557 | 0.10 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr1_-_67065797 | 0.10 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr1_+_86972244 | 0.10 |
ENSRNOT00000036907
|
Rinl
|
Ras and Rab interactor-like |
| chr19_-_37528011 | 0.10 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chrM_+_7919 | 0.09 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr3_-_40477754 | 0.09 |
ENSRNOT00000091168
|
AABR07052166.1
|
|
| chr3_+_138174054 | 0.09 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr1_+_55219773 | 0.09 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr13_-_44345735 | 0.09 |
ENSRNOT00000005006
|
Tmem163
|
transmembrane protein 163 |
| chr9_+_117795132 | 0.09 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr19_-_28751584 | 0.09 |
ENSRNOT00000079270
|
AABR07043456.1
|
|
| chr1_+_213886775 | 0.09 |
ENSRNOT00000020864
|
Pkp3
|
plakophilin 3 |
| chr10_+_56662561 | 0.09 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr3_+_150801289 | 0.09 |
ENSRNOT00000035060
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr19_-_49448072 | 0.09 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr7_+_120153184 | 0.09 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr17_-_43807540 | 0.09 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr13_+_89386023 | 0.08 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr15_+_33606124 | 0.08 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chrX_-_88321834 | 0.08 |
ENSRNOT00000020364
|
LOC100909548
|
cation-dependent mannose-6-phosphate receptor-like |
| chr20_+_1749716 | 0.08 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr3_-_161294125 | 0.08 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chr10_+_55626741 | 0.08 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chrX_+_11084317 | 0.08 |
ENSRNOT00000031808
ENSRNOT00000093745 |
RGD1565685
|
similar to RIKEN cDNA 1810030O07 |
| chr13_-_47916185 | 0.08 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr10_+_88227360 | 0.08 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr8_-_72204730 | 0.08 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr7_+_136182224 | 0.08 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr10_+_55712043 | 0.08 |
ENSRNOT00000010141
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr6_+_8886591 | 0.08 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr20_-_9291610 | 0.08 |
ENSRNOT00000000650
|
Glo1
|
glyoxalase 1 |
| chr12_+_18679789 | 0.08 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr3_-_101547478 | 0.08 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr1_-_80783898 | 0.08 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr4_-_162025090 | 0.08 |
ENSRNOT00000085887
ENSRNOT00000009904 |
Klrb1a
|
killer cell lectin-like receptor subfamily B, member 1A |
| chrM_+_9870 | 0.08 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr2_+_93758919 | 0.08 |
ENSRNOT00000077782
|
Fabp12
|
fatty acid binding protein 12 |
| chr10_+_55627025 | 0.07 |
ENSRNOT00000091016
|
Aurkb
|
aurora kinase B |
| chr16_-_75346122 | 0.07 |
ENSRNOT00000018529
|
Defa24
|
defensin alpha 24 |
| chr20_+_4966817 | 0.07 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_266141581 | 0.07 |
ENSRNOT00000078187
ENSRNOT00000051951 |
Rpe65
|
RPE65, retinoid isomerohydrolase |
| chr14_-_79464770 | 0.07 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr4_-_117575154 | 0.07 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr7_+_140608434 | 0.07 |
ENSRNOT00000001163
|
AC114446.1
|
|
| chr5_-_12526962 | 0.07 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr1_-_116153722 | 0.07 |
ENSRNOT00000041605
|
Fpr3
|
formyl peptide receptor 3 |
| chr3_+_47578384 | 0.07 |
ENSRNOT00000082769
|
Psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr12_+_41463922 | 0.07 |
ENSRNOT00000089378
|
Cfap73
|
cilia and flagella associated protein 73 |
| chr9_-_1012450 | 0.07 |
ENSRNOT00000051449
|
LOC100359951
|
ribosomal protein S20-like |
| chrX_-_64702441 | 0.07 |
ENSRNOT00000051132
|
Amer1
|
APC membrane recruitment protein 1 |
| chr4_-_30338679 | 0.07 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr4_-_159399634 | 0.07 |
ENSRNOT00000089193
|
Ndufa9
|
NADH:ubiquinone oxidoreductase subunit A9 |
| chr15_+_11298478 | 0.07 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr15_+_44411865 | 0.07 |
ENSRNOT00000017566
|
Kctd9
|
potassium channel tetramerization domain containing 9 |
| chr10_+_56662242 | 0.07 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr1_-_102741581 | 0.07 |
ENSRNOT00000016254
|
LOC691143
|
similar to Serum amyloid A-3 protein precursor |
| chr12_+_19196611 | 0.07 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr19_+_320906 | 0.07 |
ENSRNOT00000041664
|
AABR07042611.1
|
|
| chrX_+_28593405 | 0.06 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr3_-_94808861 | 0.06 |
ENSRNOT00000038464
|
Prrg4
|
proline rich and Gla domain 4 |
| chr5_-_134638150 | 0.06 |
ENSRNOT00000013480
|
Tex38
|
testis expressed 38 |
| chr20_+_13817795 | 0.06 |
ENSRNOT00000036518
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr2_+_200397967 | 0.06 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr1_-_101095594 | 0.06 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr4_+_31333970 | 0.06 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr4_+_147844658 | 0.06 |
ENSRNOT00000042673
|
H1foo
|
H1 histone family, member O, oocyte-specific |
| chr1_+_169145445 | 0.06 |
ENSRNOT00000034019
|
LOC499219
|
hypothetical protein LOC499219 |
| chr7_-_130350570 | 0.06 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr5_+_137257287 | 0.06 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chrM_+_10160 | 0.06 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr18_-_27520295 | 0.06 |
ENSRNOT00000027544
|
Gfra3
|
GDNF family receptor alpha 3 |
| chr17_+_76008807 | 0.06 |
ENSRNOT00000070895
|
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr13_-_102643223 | 0.06 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr4_-_176528110 | 0.06 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr4_-_176909075 | 0.06 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr1_-_215536980 | 0.06 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr4_-_51844331 | 0.06 |
ENSRNOT00000003593
|
Gpr37
|
G protein-coupled receptor 37 |
| chr3_-_2490392 | 0.06 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr5_+_78384444 | 0.06 |
ENSRNOT00000034978
|
LOC500475
|
similar to hypothetical protein 4933430I17 |
| chr3_-_103745236 | 0.06 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr1_+_213636093 | 0.06 |
ENSRNOT00000019642
|
Psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr8_+_99632803 | 0.06 |
ENSRNOT00000087190
|
Plscr1
|
phospholipid scramblase 1 |
| chr10_+_13836128 | 0.06 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr10_-_58608907 | 0.06 |
ENSRNOT00000010533
|
Aipl1
|
aryl hydrocarbon receptor-interacting protein-like 1 |
| chr3_-_8955538 | 0.06 |
ENSRNOT00000040570
|
LOC686066
|
similar to 60S ribosomal protein L38 |
| chr1_-_282492912 | 0.06 |
ENSRNOT00000047883
|
AABR07007130.1
|
|
| chr4_+_6827429 | 0.06 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr8_+_91820783 | 0.06 |
ENSRNOT00000043351
|
RGD1560917
|
similar to mitochondrial ribosomal protein L41 |
| chrX_+_63542191 | 0.06 |
ENSRNOT00000073955
|
Apoo
|
apolipoprotein O |
| chr12_-_19167015 | 0.06 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr9_+_98113346 | 0.06 |
ENSRNOT00000026912
|
Prlh
|
prolactin releasing hormone |
| chr15_-_30147793 | 0.06 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr6_+_2216623 | 0.06 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrY_-_1398030 | 0.06 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr20_+_44680449 | 0.06 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr7_+_29909120 | 0.05 |
ENSRNOT00000049362
|
AABR07056556.1
|
|
| chr3_-_46942966 | 0.05 |
ENSRNOT00000087439
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr13_+_67545430 | 0.05 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr9_+_94562839 | 0.05 |
ENSRNOT00000022308
|
RGD1311447
|
LOC363276 |
| chr1_+_105285419 | 0.05 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr10_+_56445647 | 0.05 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr5_+_128450680 | 0.05 |
ENSRNOT00000010700
|
Txndc12
|
thioredoxin domain containing 12 |
| chr19_+_37873515 | 0.05 |
ENSRNOT00000026163
ENSRNOT00000089921 |
Pskh1
|
protein serine kinase H1 |
| chr4_-_170912629 | 0.05 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr14_-_21299068 | 0.05 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr1_-_216663720 | 0.05 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr4_+_70977556 | 0.05 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr7_+_72924799 | 0.05 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr19_-_41349681 | 0.05 |
ENSRNOT00000080694
ENSRNOT00000059147 |
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr3_-_148493225 | 0.05 |
ENSRNOT00000012141
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr1_+_22332090 | 0.05 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr8_-_22785671 | 0.05 |
ENSRNOT00000045384
|
Spc24
|
SPC24, NDC80 kinetochore complex component |
| chr3_+_16846412 | 0.05 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr11_-_17684903 | 0.05 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr7_-_143324536 | 0.05 |
ENSRNOT00000011644
|
Krt5
|
keratin 5 |
| chrM_+_8599 | 0.05 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr10_+_74413989 | 0.05 |
ENSRNOT00000036098
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_-_165606375 | 0.05 |
ENSRNOT00000024290
|
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr7_-_28711761 | 0.05 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr10_-_83898527 | 0.05 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr13_-_27192592 | 0.05 |
ENSRNOT00000040021
|
Serpinb3
|
serpin family B member 3 |
| chr16_-_31301880 | 0.05 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr1_+_196095214 | 0.05 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chr10_-_44147035 | 0.05 |
ENSRNOT00000041113
|
Olr1424
|
olfactory receptor 1424 |
| chr11_+_70056624 | 0.05 |
ENSRNOT00000002447
|
AC133403.1
|
|
| chr3_+_94530586 | 0.05 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr11_+_36851038 | 0.05 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr10_+_103266296 | 0.05 |
ENSRNOT00000038852
|
Dnai2
|
dynein, axonemal, intermediate chain 2 |
| chr2_-_27287605 | 0.05 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr14_+_22375955 | 0.05 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr8_-_96516975 | 0.05 |
ENSRNOT00000067996
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr1_-_220136470 | 0.05 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chr10_+_71217966 | 0.05 |
ENSRNOT00000076192
|
Hnf1b
|
HNF1 homeobox B |
| chr16_-_6669045 | 0.05 |
ENSRNOT00000067639
|
Prkcd
|
protein kinase C, delta |
| chr7_+_18683553 | 0.05 |
ENSRNOT00000009425
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr16_+_54332660 | 0.05 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr8_+_99625545 | 0.05 |
ENSRNOT00000010689
ENSRNOT00000056727 |
Plscr1
|
phospholipid scramblase 1 |
| chr15_-_32888095 | 0.05 |
ENSRNOT00000012233
|
Dad1
|
defender against cell death 1 |
| chr2_-_30246010 | 0.05 |
ENSRNOT00000023900
|
Mccc2
|
methylcrotonoyl-CoA carboxylase 2 |
| chr7_+_107413691 | 0.05 |
ENSRNOT00000007587
|
Phf20l1
|
PHD finger protein 20-like protein 1 |
| chr9_-_73948583 | 0.05 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr16_+_37177443 | 0.05 |
ENSRNOT00000014083
|
Cep44
|
centrosomal protein 44 |
| chr10_+_13230158 | 0.05 |
ENSRNOT00000075648
|
Csap1
|
common salivary protein 1 |
| chr4_-_158705885 | 0.05 |
ENSRNOT00000087115
ENSRNOT00000026685 |
Ntf3
|
neurotrophin 3 |
| chr10_-_72196437 | 0.05 |
ENSRNOT00000029769
|
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr5_-_17061361 | 0.05 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr14_-_6533524 | 0.05 |
ENSRNOT00000079795
|
Abcg3l1
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 1 |
| chr8_+_122076759 | 0.05 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr1_-_224698514 | 0.05 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chrX_-_77675487 | 0.05 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr5_-_174921 | 0.05 |
ENSRNOT00000056239
|
Tipinl1
|
TIMELESS interacting protein like 1 |
| chr19_+_37282018 | 0.05 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr5_-_134927235 | 0.05 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr12_-_21746236 | 0.05 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr1_+_227892956 | 0.05 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr2_-_187706300 | 0.05 |
ENSRNOT00000092349
ENSRNOT00000026414 |
Tmem79
|
transmembrane protein 79 |
| chr1_+_102924059 | 0.05 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr14_-_84662143 | 0.04 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr10_-_87459652 | 0.04 |
ENSRNOT00000018804
|
Krt40
|
keratin 40 |
| chr2_-_197991574 | 0.04 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr10_-_14324170 | 0.04 |
ENSRNOT00000035513
ENSRNOT00000090587 |
Hn1l
|
hematological and neurological expressed 1-like |
| chr2_-_197991198 | 0.04 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr6_-_91581262 | 0.04 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx5_Dlx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0036446 | myofibroblast differentiation(GO:0036446) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.1 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0035565 | regulation of pronephros size(GO:0035565) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:2000373 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.0 | GO:0042335 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.0 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.0 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.0 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.0 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.0 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.0 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.0 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) phenotypic switching(GO:0036166) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070821 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.1 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.7 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |