Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
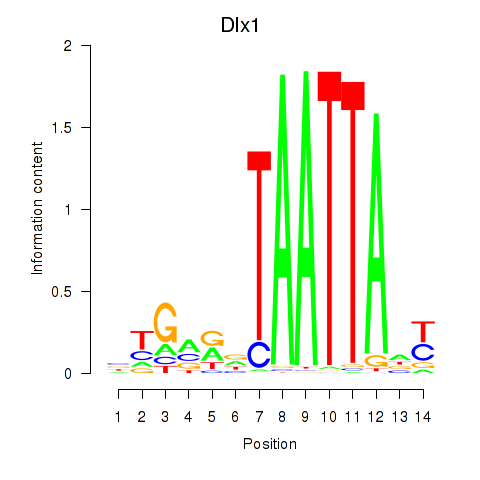
Results for Dlx1
Z-value: 0.62
Transcription factors associated with Dlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dlx1
|
ENSRNOG00000001520 | distal-less homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx1 | rn6_v1_chr3_+_58164931_58164931 | -0.65 | 2.3e-01 | Click! |
Activity profile of Dlx1 motif
Sorted Z-values of Dlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_104155775 | 0.54 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chrM_+_9870 | 0.37 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr5_+_6373583 | 0.36 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr2_+_187447501 | 0.34 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr7_-_145062956 | 0.24 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr10_-_44746549 | 0.24 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chrM_+_3904 | 0.22 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr10_+_55627025 | 0.21 |
ENSRNOT00000091016
|
Aurkb
|
aurora kinase B |
| chrX_+_20520034 | 0.21 |
ENSRNOT00000093170
|
FAM120C
|
family with sequence similarity 120C |
| chr2_-_219262901 | 0.20 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr1_+_196095214 | 0.17 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chr7_+_140608434 | 0.17 |
ENSRNOT00000001163
|
AC114446.1
|
|
| chrM_+_9451 | 0.16 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrM_+_11736 | 0.16 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrX_+_84064427 | 0.16 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr13_+_96303703 | 0.16 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr2_+_58724855 | 0.16 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr20_+_4593389 | 0.15 |
ENSRNOT00000001174
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr7_-_76035096 | 0.15 |
ENSRNOT00000072255
|
AABR07057530.1
|
|
| chr15_-_44442875 | 0.15 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr8_-_96547568 | 0.14 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr1_+_211582077 | 0.14 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr1_-_48891130 | 0.14 |
ENSRNOT00000083884
|
AC135026.1
|
|
| chr8_-_17696989 | 0.14 |
ENSRNOT00000083752
|
AABR07069336.1
|
|
| chr20_+_46044892 | 0.13 |
ENSRNOT00000057187
|
Ak9
|
adenylate kinase 9 |
| chr3_-_40477754 | 0.13 |
ENSRNOT00000091168
|
AABR07052166.1
|
|
| chr1_-_198104109 | 0.13 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr15_-_42794279 | 0.13 |
ENSRNOT00000023385
|
Ephx2
|
epoxide hydrolase 2 |
| chr5_-_17061837 | 0.13 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr4_+_148782479 | 0.13 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr4_+_69386698 | 0.13 |
ENSRNOT00000091655
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr2_-_147819335 | 0.13 |
ENSRNOT00000057909
|
Ankub1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr2_+_178117466 | 0.13 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr4_+_70977556 | 0.13 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chrX_-_70978952 | 0.13 |
ENSRNOT00000076910
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr10_+_55626741 | 0.13 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr5_-_17061361 | 0.13 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chrX_-_106607352 | 0.12 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr4_-_176528110 | 0.12 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr6_-_91581262 | 0.12 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr11_-_13999862 | 0.12 |
ENSRNOT00000045835
|
Lipi
|
lipase I |
| chr10_-_66229311 | 0.12 |
ENSRNOT00000016897
|
Lgals5
|
lectin, galactose binding, soluble 5 |
| chr2_-_256154584 | 0.12 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr15_-_59215803 | 0.12 |
ENSRNOT00000032301
|
Lacc1
|
laccase domain containing 1 |
| chr1_+_80777014 | 0.11 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr13_-_83457888 | 0.11 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr5_-_134927235 | 0.11 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr10_+_56445647 | 0.11 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr19_+_15081590 | 0.11 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr15_+_36809361 | 0.11 |
ENSRNOT00000076667
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr7_-_54855557 | 0.11 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr10_-_34242985 | 0.11 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr18_-_6587080 | 0.11 |
ENSRNOT00000040815
|
LOC103694404
|
60S ribosomal protein L39 |
| chr3_-_4341771 | 0.11 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr1_+_86972244 | 0.11 |
ENSRNOT00000036907
|
Rinl
|
Ras and Rab interactor-like |
| chr1_+_185863043 | 0.11 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr15_+_11298478 | 0.11 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr15_+_87722221 | 0.10 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr17_+_11683862 | 0.10 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr1_+_98414226 | 0.10 |
ENSRNOT00000090785
|
Siglecl1
|
SIGLEC family like 1 |
| chr10_-_13004441 | 0.10 |
ENSRNOT00000004848
|
Cldn9
|
claudin 9 |
| chr4_+_31333970 | 0.10 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr8_+_70994563 | 0.10 |
ENSRNOT00000051504
ENSRNOT00000077163 |
Spg21
|
spastic paraplegia 21 homolog (human) |
| chr3_+_172374957 | 0.10 |
ENSRNOT00000035916
|
Gnas
|
GNAS complex locus |
| chr3_+_94530586 | 0.10 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr20_+_8484311 | 0.10 |
ENSRNOT00000050041
|
LOC100364116
|
ribosomal protein S20-like |
| chr17_+_24416651 | 0.10 |
ENSRNOT00000024458
|
Cd83
|
CD83 molecule |
| chr2_-_149444548 | 0.10 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr11_+_38727048 | 0.10 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr4_-_51844331 | 0.09 |
ENSRNOT00000003593
|
Gpr37
|
G protein-coupled receptor 37 |
| chr3_-_143192408 | 0.09 |
ENSRNOT00000006885
|
LOC689081
|
similar to cystatin E2 |
| chr12_+_41463922 | 0.09 |
ENSRNOT00000089378
|
Cfap73
|
cilia and flagella associated protein 73 |
| chr11_-_71136673 | 0.09 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr13_+_89386023 | 0.09 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr2_-_27287605 | 0.09 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr20_+_2004052 | 0.09 |
ENSRNOT00000001008
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr1_-_102741581 | 0.09 |
ENSRNOT00000016254
|
LOC691143
|
similar to Serum amyloid A-3 protein precursor |
| chr5_+_36566783 | 0.09 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr4_-_176909075 | 0.09 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr4_-_164453171 | 0.09 |
ENSRNOT00000077539
ENSRNOT00000083610 ENSRNOT00000079975 |
Ly49s6
|
Ly49 stimulatory receptor 6 |
| chr8_+_76426335 | 0.09 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr1_-_78180216 | 0.09 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr1_+_252409268 | 0.09 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chrM_+_7006 | 0.08 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chrM_+_7919 | 0.08 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chrX_+_118197217 | 0.08 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr5_-_12172009 | 0.08 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr18_+_16330615 | 0.08 |
ENSRNOT00000049689
|
RGD1559877
|
similar to 60S ribosomal protein L29 (P23) |
| chr3_-_90751055 | 0.08 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr2_+_153803349 | 0.08 |
ENSRNOT00000088565
|
Mme
|
membrane metallo-endopeptidase |
| chrM_+_14136 | 0.08 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr8_+_2604962 | 0.08 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr8_-_109560747 | 0.08 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_-_29949603 | 0.08 |
ENSRNOT00000009419
|
LOC103692829
|
60S ribosomal protein L9 pseudogene |
| chr9_+_95161157 | 0.08 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr9_-_14550625 | 0.08 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr18_+_87637608 | 0.08 |
ENSRNOT00000070851
|
LOC102550729
|
zinc finger protein 120-like |
| chr11_-_29710849 | 0.08 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr1_-_57518458 | 0.08 |
ENSRNOT00000002040
|
Pdcd2
|
programmed cell death 2 |
| chr11_+_36851038 | 0.08 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr20_-_27117663 | 0.08 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr2_+_187951344 | 0.08 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr1_+_207508414 | 0.08 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chrM_+_2740 | 0.08 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chrM_+_8599 | 0.08 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr10_+_88227360 | 0.08 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr2_+_88344527 | 0.07 |
ENSRNOT00000059467
|
RGD1565641
|
RGD1565641 |
| chrX_+_14019961 | 0.07 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr7_+_144052061 | 0.07 |
ENSRNOT00000020103
ENSRNOT00000091378 |
Amhr2
|
anti-Mullerian hormone receptor type 2 |
| chr14_-_18704059 | 0.07 |
ENSRNOT00000081455
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr3_-_114251647 | 0.07 |
ENSRNOT00000024245
|
Duoxa1
|
dual oxidase maturation factor 1 |
| chr3_-_148493225 | 0.07 |
ENSRNOT00000012141
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr9_+_66058047 | 0.07 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr1_+_15834779 | 0.07 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr1_-_276228574 | 0.07 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr2_+_66940057 | 0.07 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr3_-_57646572 | 0.07 |
ENSRNOT00000076202
|
Mettl8
|
methyltransferase like 8 |
| chrX_+_28593405 | 0.07 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr7_-_143324536 | 0.07 |
ENSRNOT00000011644
|
Krt5
|
keratin 5 |
| chr4_+_2053712 | 0.07 |
ENSRNOT00000045086
|
Rnf32
|
ring finger protein 32 |
| chr17_+_21600432 | 0.07 |
ENSRNOT00000060340
|
RGD1562963
|
similar to chromosome 6 open reading frame 52 |
| chr3_+_47578384 | 0.07 |
ENSRNOT00000082769
|
Psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr5_-_127983515 | 0.07 |
ENSRNOT00000043054
|
Fam159a
|
family with sequence similarity 159, member A |
| chr18_-_40452456 | 0.07 |
ENSRNOT00000004747
|
Mospd4
|
motile sperm domain containing 4 |
| chr18_+_81821127 | 0.07 |
ENSRNOT00000058199
|
Fbxo15
|
F-box protein 15 |
| chr5_-_157268903 | 0.07 |
ENSRNOT00000022716
|
Pla2g5
|
phospholipase A2, group V |
| chr2_+_127489771 | 0.07 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr14_+_3204390 | 0.07 |
ENSRNOT00000088078
|
Glmn
|
glomulin, FKBP associated protein |
| chr19_+_15081158 | 0.07 |
ENSRNOT00000074070
|
Ces1f
|
carboxylesterase 1F |
| chr1_+_61268248 | 0.07 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr1_+_116604550 | 0.07 |
ENSRNOT00000083587
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr1_-_71373605 | 0.07 |
ENSRNOT00000034854
|
Galp
|
galanin-like peptide |
| chr3_+_154437571 | 0.07 |
ENSRNOT00000019331
|
AABR07054456.1
|
|
| chr4_+_6827429 | 0.07 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr11_+_88095170 | 0.07 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr2_+_145174876 | 0.06 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr11_-_54545230 | 0.06 |
ENSRNOT00000002676
|
Retnla
|
resistin like alpha |
| chr3_-_153114520 | 0.06 |
ENSRNOT00000008254
|
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr14_+_108415068 | 0.06 |
ENSRNOT00000083763
|
Pus10
|
pseudouridylate synthase 10 |
| chr2_-_27296777 | 0.06 |
ENSRNOT00000080748
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chrX_-_37705263 | 0.06 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr9_-_30251388 | 0.06 |
ENSRNOT00000035033
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr14_+_18983853 | 0.06 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr1_-_282492912 | 0.06 |
ENSRNOT00000047883
|
AABR07007130.1
|
|
| chr7_-_28711761 | 0.06 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr2_+_202200797 | 0.06 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chrX_-_45284341 | 0.06 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr1_-_38538987 | 0.06 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr18_+_45023932 | 0.06 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr6_+_93385457 | 0.06 |
ENSRNOT00000010300
|
Actr10
|
actin-related protein 10 homolog |
| chr5_+_126668689 | 0.06 |
ENSRNOT00000036072
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr20_+_48503973 | 0.06 |
ENSRNOT00000064081
|
Cdc40
|
cell division cycle 40 |
| chr5_+_78384444 | 0.06 |
ENSRNOT00000034978
|
LOC500475
|
similar to hypothetical protein 4933430I17 |
| chr7_-_55604403 | 0.06 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr4_+_181103774 | 0.06 |
ENSRNOT00000084207
ENSRNOT00000055473 ENSRNOT00000077619 |
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chrX_+_45213228 | 0.06 |
ENSRNOT00000088538
|
LOC103690319
|
odorant-binding protein |
| chr8_-_133128290 | 0.06 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr19_-_49448072 | 0.06 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr6_+_76349362 | 0.06 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr1_-_94404211 | 0.06 |
ENSRNOT00000019463
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr16_+_71889235 | 0.06 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr10_+_91126689 | 0.06 |
ENSRNOT00000004046
|
Nmt1
|
N-myristoyltransferase 1 |
| chr10_+_74413989 | 0.06 |
ENSRNOT00000036098
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_52477575 | 0.06 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr10_-_56506446 | 0.06 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr10_-_44147035 | 0.06 |
ENSRNOT00000041113
|
Olr1424
|
olfactory receptor 1424 |
| chr4_-_89695928 | 0.06 |
ENSRNOT00000039316
|
Gprin3
|
GPRIN family member 3 |
| chr8_-_21384131 | 0.06 |
ENSRNOT00000071010
|
Zfp560
|
zinc finger protein 560 |
| chr1_+_222861777 | 0.06 |
ENSRNOT00000090872
|
Pla2g16
|
phospholipase A2, group XVI |
| chr14_-_6533524 | 0.06 |
ENSRNOT00000079795
|
Abcg3l1
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 1 |
| chr1_-_224698514 | 0.06 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr1_+_61374379 | 0.06 |
ENSRNOT00000015343
|
Zfp53
|
zinc finger protein 53 |
| chr7_+_29909120 | 0.06 |
ENSRNOT00000049362
|
AABR07056556.1
|
|
| chr13_-_47916185 | 0.06 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr8_+_22559098 | 0.06 |
ENSRNOT00000041091
|
LOC691141
|
hypothetical protein LOC691141 |
| chr15_+_33885106 | 0.06 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr10_+_39850818 | 0.06 |
ENSRNOT00000012671
|
Fnip1
|
folliculin interacting protein 1 |
| chr18_-_36579403 | 0.06 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr10_-_51778939 | 0.06 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr2_+_184243976 | 0.06 |
ENSRNOT00000042146
|
Dear
|
dual endothelin 1, angiotensin II receptor |
| chr17_+_45467015 | 0.06 |
ENSRNOT00000081408
|
Gpx5
|
glutathione peroxidase 5 |
| chr12_-_17186679 | 0.06 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr5_+_50381244 | 0.06 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr7_-_107203897 | 0.06 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr13_+_111890894 | 0.06 |
ENSRNOT00000007341
|
LOC100125367
|
hypothetical protein LOC100125367 |
| chr2_+_243425007 | 0.05 |
ENSRNOT00000082894
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr7_+_15422479 | 0.05 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr18_-_26656879 | 0.05 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr8_+_122076759 | 0.05 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr12_+_46042413 | 0.05 |
ENSRNOT00000046882
|
Ccdc60
|
coiled-coil domain containing 60 |
| chr4_+_112662609 | 0.05 |
ENSRNOT00000009104
|
Gcfc2
|
GC-rich sequence DNA-binding factor 2 |
| chr16_+_72010106 | 0.05 |
ENSRNOT00000058330
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr8_-_72204730 | 0.05 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr3_-_2309179 | 0.05 |
ENSRNOT00000012375
|
Noxa1
|
NADPH oxidase activator 1 |
| chr3_+_10036461 | 0.05 |
ENSRNOT00000073731
|
Qrfp
|
pyroglutamylated RFamide peptide |
| chrX_-_121731543 | 0.05 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr2_-_90568486 | 0.05 |
ENSRNOT00000059380
|
LOC100910852
|
uncharacterized LOC100910852 |
| chr3_-_133131192 | 0.05 |
ENSRNOT00000055630
|
Tasp1
|
taspase 1 |
| chr9_-_121972055 | 0.05 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr13_-_45127815 | 0.05 |
ENSRNOT00000005127
|
Dars
|
aspartyl-tRNA synthetase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.0 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 0.0 | 0.1 | GO:0009216 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.0 | 0.1 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0000962 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.0 | 0.2 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:1900222 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.0 | 1.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.0 | GO:0072702 | myofibroblast differentiation(GO:0036446) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.1 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.0 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.0 | 0.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.0 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.0 | 0.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.0 | 0.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.0 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |